 |
PDBsum entry 1m1t

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.9
- acetyl-CoA C-acetyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Mevalonate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 acetyl-CoA = acetoacetyl-CoA + CoA
|
 |
 |
 |
 |
 |
 2
×
acetyl-CoA
2
×
acetyl-CoA
|
=
|
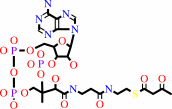 acetoacetyl-CoA
acetoacetyl-CoA
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:15543-15556
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
The catalytic cycle of biosynthetic thiolase: a conformational journey of an acetyl group through four binding modes and two oxyanion holes.
|
|
P.Kursula,
J.Ojala,
A.M.Lambeir,
R.K.Wierenga.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Biosynthetic thiolase catalyzes the formation of acetoacetyl-CoA from two
molecules of acetyl-CoA. This is a key step in the synthesis of many biological
compounds, including steroid hormones and ketone bodies. The thiolase reaction
involves two chemically distinct steps; during acyl transfer, an acetyl group is
transferred from acetyl-CoA to Cys89, and in the Claisen condensation step, this
acetyl group is further transferred to a second molecule of acetyl-CoA,
generating acetoacetyl-CoA. Here, new crystallographic data for Zoogloea
ramigera biosynthetic thiolase are presented, covering all intermediates of the
thiolase catalytic cycle. The high-resolution structures indicate that the
acetyl group goes through four conformations while being transferred from
acetyl-CoA via the acetylated enzyme to acetoacetyl-CoA. This transfer is
catalyzed in a rigid cavity lined by mostly hydrophobic side chains, in addition
to the catalytic residues Cys89, His348, and Cys378. The structures highlight
the importance of an oxyanion hole formed by a water molecule and His348 in
stabilizing the negative charge on the thioester oxygen atom of acetyl-CoA at
two different steps of the reaction cycle. Another oxyanion hole, composed of
the main chain nitrogen atoms of Cys89 and Gly380, complements a negative charge
of the thioester oxygen anion of the acetylated intermediate, stabilizing the
tetrahedral transition state of the Claisen condensation step. The reactivity of
the active site may be modulated by hydrogen bonding networks extending from the
active site toward the back of the molecule.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
W.Yang,
D.Di Vizio,
M.Kirchner,
H.Steen,
and
M.R.Freeman
(2010).
Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes.
|
| |
Mol Cell Proteomics,
9,
54-70.
|
 |
|
|
|
|
 |
G.Meriläinen,
W.Schmitz,
R.K.Wierenga,
and
P.Kursula
(2008).
The sulfur atoms of the substrate CoA and the catalytic cysteine are required for a productive mode of substrate binding in bacterial biosynthetic thiolase, a thioester-dependent enzyme.
|
| |
FEBS J,
275,
6136-6148.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Boekhorst,
and
B.Snel
(2007).
Identification of homologs in insignificant blast hits by exploiting extrinsic gene properties.
|
| |
BMC Bioinformatics,
8,
356.
|
 |
|
|
|
|
 |
A.M.Haapalainen,
G.Meriläinen,
and
R.K.Wierenga
(2006).
The thiolase superfamily: condensing enzymes with diverse reaction specificities.
|
| |
Trends Biochem Sci,
31,
64-71.
|
 |
|
|
|
|
 |
Y.Meng,
and
J.Li
(2006).
Cloning, expression and characterization of a thiolase gene from Clostridium pasteurianum.
|
| |
Biotechnol Lett,
28,
1227-1232.
|
 |
|
|
|
|
 |
F.Schroeder,
H.Huang,
H.A.Hostetler,
A.D.Petrescu,
R.Hertz,
J.Bar-Tana,
and
A.B.Kier
(2005).
Stability of fatty acyl-coenzyme A thioester ligands of hepatocyte nuclear factor-4alpha and peroxisome proliferator-activated receptor-alpha.
|
| |
Lipids,
40,
559-568.
|
 |
|
|
|
|
 |
P.Y.Wu,
M.Hanlon,
M.Eddins,
C.Tsui,
R.S.Rogers,
J.P.Jensen,
M.J.Matunis,
A.M.Weissman,
A.M.Weisman,
A.M.Weissman,
C.Wolberger,
C.P.Wolberger,
and
C.M.Pickart
(2003).
A conserved catalytic residue in the ubiquitin-conjugating enzyme family.
|
| |
EMBO J,
22,
5241-5250.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

