 |
PDBsum entry 1la2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Structural analysis of saccharomyces cerevisiae myo-inositol phosphate synthase
|
|
Structure:
|
 |
Myo-inositol-1-phosphate synthase. Chain: a, b, c, d. Synonym: mi-1-p synthase, ips. Engineered: yes
|
|
Source:
|
 |
Saccharomyces cerevisiae. Baker's yeast. Organism_taxid: 4932. Gene: ino1. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.65Å
|
R-factor:
|
0.224
|
R-free:
|
0.280
|
|
|
Authors:
|
 |
R.Kniewel,J.A.Buglino,V.Shen,T.Chadna,A.Beckwith,C.D.Lima,S.K.Burley, New York Sgx Research Center For Structural Genomics (Nysgxrc)
|
|
Key ref:
|
 |
R.Kniewel
et al.
(2002).
Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase.
J Struct Funct Genomics,
2,
129-134.
PubMed id:
|
 |
|
Date:
|
 |
|
27-Mar-02
|
Release date:
|
10-Apr-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P11986
(INO1_YEAST) -
Inositol-3-phosphate synthase from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
533 a.a.
514 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.5.1.4
- inositol-3-phosphate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose 6-phosphate = 1D-myo-inositol 3-phosphate
|
 |
 |
 |
 |
 |
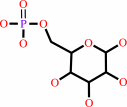 D-glucose 6-phosphate
D-glucose 6-phosphate
|
=
|
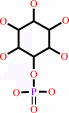 1D-myo-inositol 3-phosphate
1D-myo-inositol 3-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
NAD(+)
|
 |
 |
 |
 |
 |
NAD(+)
Bound ligand (Het Group name =
NAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Struct Funct Genomics
2:129-134
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase.
|
|
R.Kniewel,
J.A.Buglino,
V.Shen,
T.Chadha,
A.Beckwith,
C.D.Lima.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The New York Structural Genomics Research Consortium has targeted highly
conserved but uncharacterized enzyme families for structure determination. As
part of this effort, the 2.65-A crystal structure has been determined for
Saccharomyces cerevisiae myo-inositol 1-phosphate synthase (MIP), an essential
enzyme that catalyzes critical steps in inositol biosynthesis. The structure
determination of four independent monomers in the asymmetric unit (240 kDa)
reveals atomic details and residue composition for the partially closed
NAD-containing active sites in apo-configuration. The structure further reveals
extensive interactions involved in tetrameric assembly of the enzyme complex.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.M.Flatt,
and
T.Mahmud
(2007).
Biosynthesis of aminocyclitol-aminoglycoside antibiotics and related compounds.
|
| |
Nat Prod Rep,
24,
358-392.
|
 |
|
|
|
|
 |
M.Majee,
S.Maitra,
K.G.Dastidar,
S.Pattnaik,
A.Chatterjee,
N.C.Hait,
K.P.Das,
and
A.L.Majumder
(2004).
A novel salt-tolerant L-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice: molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype.
|
| |
J Biol Chem,
279,
28539-28552.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

