
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 511 a.a.
511 a.a.
|
 |

|

|

|

|

|

|

|

 388 a.a.
388 a.a.
|
 |

|

|

|

|

|

|

|

 167 a.a.
167 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Methane monooxygenase hydroxylase, form ii cocrystallized with dibromomethane
|
|
Structure:
|
 |
Methane monooxygenase component a, alpha chain. Chain: a, b. Synonym: hydroxylase alpha subunit. Methane monooxygenase component a, beta chain. Chain: c, d. Synonym: hydroxylase beta subunit, methane monooxygenase a beta chain. Methane monooxygenase component a, gamma chain. Chain: e, f.
|
|
Source:
|
 |
Methylococcus capsulatus. Organism_taxid: 414. Organism_taxid: 414
|
|
Biol. unit:
|
 |
Hexamer (from
)
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.201
|
R-free:
|
0.250
|
|
|
Authors:
|
 |
D.A.Whittington,A.C.Rosenzweig,C.A.Frederick,S.J.Lippard
|
Key ref:

|
 |
D.A.Whittington
et al.
(2001).
Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry,
40,
3476-3482.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
03-Oct-00
|
Release date:
|
27-Apr-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P22869
(MEMA_METCA) -
Methane monooxygenase component A alpha chain from Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
527 a.a.
511 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.1.14.13.25
- methane monooxygenase (soluble).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
methane + NADH + O2 + H+ = methanol + NAD+ + H2O
|
|
2.
|
methane + NADPH + O2 + H+ = methanol + NADP+ + H2O
|
|
 |
 |
 |
 |
 |
methane
|
+
|
 NADH
NADH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
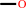 methanol
methanol
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
methane
|
+
|
 NADPH
NADPH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
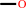 methanol
methanol
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
40:3476-3482
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
|
|
D.A.Whittington,
A.C.Rosenzweig,
C.A.Frederick,
S.J.Lippard.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
To investigate the role of protein cavities in facilitating movement of the
substrates, methane and dioxygen, in the soluble methane monooxygenase
hydroxylase (MMOH), we determined the X-ray structures of MMOH from
Methylococcus capsulatus (Bath) cocrystallized with dibromomethane or
iodoethane, or by using crystals pressurized with xenon gas. The halogenated
alkanes bind in two cavities within the alpha-subunit that extend from one
surface of the protein to the buried dinuclear iron active site. Two additional
binding sites were located in the beta-subunit. Pressurization of two crystal
forms of MMOH with xenon resulted in the identification of six binding sites
located exclusively in the alpha-subunit. These results indicate that
hydrophobic species bind preferentially in preexisting cavities in MMOH and
support the hypothesis that such cavities may play a functional role in
sequestering and enhancing the availability of the physiological substrates for
reaction at the active site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.J.Lee,
M.S.McCormick,
S.J.Lippard,
and
U.S.Cho
(2013).
Control of substrate access to the active site in methane monooxygenase.
|
| |
Nature,
494,
380-384.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Friedle,
E.Reisner,
and
S.J.Lippard
(2010).
Current challenges of modeling diiron enzyme active sites for dioxygen activation by biomimetic synthetic complexes.
|
| |
Chem Soc Rev,
39,
2768-2779.
|
 |
|
|
|
|
 |
L.Chen,
A.Y.Lyubimov,
L.Brammer,
A.Vrielink,
and
N.S.Sampson
(2008).
The binding and release of oxygen and hydrogen peroxide are directed by a hydrophobic tunnel in cholesterol oxidase.
|
| |
Biochemistry,
47,
5368-5377.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.J.Johnson,
J.Cohen,
R.W.Welford,
A.R.Pearson,
K.Schulten,
J.P.Klinman,
and
C.M.Wilmot
(2007).
Exploring molecular oxygen pathways in Hansenula polymorpha copper-containing amine oxidase.
|
| |
J Biol Chem,
282,
17767-17776.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.H.Halsey,
L.A.Sayavedra-Soto,
P.J.Bottomley,
and
D.J.Arp
(2006).
Site-directed amino acid substitutions in the hydroxylase alpha subunit of butane monooxygenase from Pseudomonas butanovora: Implications for substrates knocking at the gate.
|
| |
J Bacteriol,
188,
4962-4969.
|
 |
|
|
|
|
 |
L.J.Murray,
R.García-Serres,
S.Naik,
B.H.Huynh,
and
S.J.Lippard
(2006).
Dioxygen activation at non-heme diiron centers: characterization of intermediates in a mutant form of toluene/o-xylene monooxygenase hydroxylase.
|
| |
J Am Chem Soc,
128,
7458-7459.
|
 |
|
|
|
|
 |
M.H.Sazinsky,
P.W.Dunten,
M.S.McCormick,
A.DiDonato,
and
S.J.Lippard
(2006).
X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase.
|
| |
Biochemistry,
45,
15392-15404.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.S.McCormick,
M.H.Sazinsky,
K.L.Condon,
and
S.J.Lippard
(2006).
X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior.
|
| |
J Am Chem Soc,
128,
15108-15110.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Dalton
(2005).
The Leeuwenhoek Lecture 2000 the natural and unnatural history of methane-oxidizing bacteria.
|
| |
Philos Trans R Soc Lond B Biol Sci,
360,
1207-1222.
|
 |
|
|
|
|
 |
D.Dantsker,
U.Samuni,
Y.Ouellet,
B.A.Wittenberg,
J.B.Wittenberg,
M.Milani,
M.Bolognesi,
M.Guertin,
and
J.M.Friedman
(2004).
Viscosity-dependent relaxation significantly modulates the kinetics of CO recombination in the truncated hemoglobin TrHbN from Mycobacterium tuberculosis.
|
| |
J Biol Chem,
279,
38844-38853.
|
 |
|
|
|
|
 |
I.Moudrakovski,
D.V.Soldatov,
J.A.Ripmeester,
D.N.Sears,
and
C.J.Jameson
(2004).
Xe NMR lineshapes in channels of peptide molecular crystals.
|
| |
Proc Natl Acad Sci U S A,
101,
17924-17929.
|
 |
|
|
|
|
 |
L.P.Wackett,
A.G.Dodge,
and
L.B.Ellis
(2004).
Microbial genomics and the periodic table.
|
| |
Appl Environ Microbiol,
70,
647-655.
|
 |
|
|
|
|
 |
M.H.Sazinsky,
J.Bard,
A.Di Donato,
and
S.J.Lippard
(2004).
Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. Insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases.
|
| |
J Biol Chem,
279,
30600-30610.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Radding,
and
G.N.Phillips
(2004).
Kinetic proofreading by the cavity system of myoglobin: protection from poisoning.
|
| |
Bioessays,
26,
422-433.
|
 |
|
|
|
|
 |
D.A.Kopp,
E.A.Berg,
C.E.Costello,
and
S.J.Lippard
(2003).
Structural features of covalently cross-linked hydroxylase and reductase proteins of soluble methane monooxygenase as revealed by mass spectrometric analysis.
|
| |
J Biol Chem,
278,
20939-20945.
|
 |
|
|
|
|
 |
U.Samuni,
D.Dantsker,
A.Ray,
J.B.Wittenberg,
B.A.Wittenberg,
S.Dewilde,
L.Moens,
Y.Ouellet,
M.Guertin,
and
J.M.Friedman
(2003).
Kinetic modulation in carbonmonoxy derivatives of truncated hemoglobins: the role of distal heme pocket residues and extended apolar tunnel.
|
| |
J Biol Chem,
278,
27241-27250.
|
 |
|
|
|
|
 |
C.M.Wilmot,
and
A.R.Pearson
(2002).
Cryocrystallography of metalloprotein reaction intermediates.
|
| |
Curr Opin Chem Biol,
6,
202-207.
|
 |
|
|
|
|
 |
D.A.Kopp,
and
S.J.Lippard
(2002).
Soluble methane monooxygenase: activation of dioxygen and methane.
|
| |
Curr Opin Chem Biol,
6,
568-576.
|
 |
|
|
|
|
 |
K.H.Mitchell,
J.M.Studts,
and
B.G.Fox
(2002).
Combined participation of hydroxylase active site residues and effector protein binding in a para to ortho modulation of toluene 4-monooxygenase regiospecificity.
|
| |
Biochemistry,
41,
3176-3188.
|
 |
|
|
|
|
 |
M.J.Ryle,
and
R.P.Hausinger
(2002).
Non-heme iron oxygenases.
|
| |
Curr Opin Chem Biol,
6,
193-201.
|
 |
|
|
|
|
 |
M.Merkx,
D.A.Kopp,
M.H.Sazinsky,
J.L.Blazyk,
J.Müller,
and
S.J.Lippard
(2001).
Dioxygen Activation and Methane Hydroxylation by Soluble Methane Monooxygenase: A Tale of Two Irons and Three Proteins A list of abbreviations can be found in Section 7.
|
| |
Angew Chem Int Ed Engl,
40,
2782-2807.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

