 |
PDBsum entry 1csc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxo-acid-lyase
|
PDB id
|
|

|

|
1csc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.3.1
- citrate (Si)-synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Citric acid cycle
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxaloacetate + acetyl-CoA + H2O = citrate + CoA + H+
|
 |
 |
 |
 |
 |
oxaloacetate
Bound ligand (Het Group name = )
matches with 94.34% similarity
|
+
|
 acetyl-CoA
acetyl-CoA
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
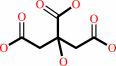 citrate
citrate
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
30:6024-6031
(1991)
|
|
PubMed id:
|
|
|
|

|
| |
|
1.9-A structures of ternary complexes of citrate synthase with D- and L-malate: mechanistic implications.
|
|
M.Karpusas,
D.Holland,
S.J.Remington.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structures of four isomorphous crystals of ternary complexes of chicken
heart citrate synthase with D- or L-malate and acetyl coenzyme A or
carboxymethyl coenzyme A have been determined by X-ray crystallography and fully
refined at 1.9-A resolution. The structures show that both L-malate and D-malate
bind in a very similar way in the presence of acetylCoA and that the enzyme
conformation is "closed". Hydrogen bond geometry is suggested to account for the
difference in binding constants of the two stereoisomers. The structures suggest
that steric hindrance can account for the observation that proton exchange of
acetyl coenzyme A with solvent is catalyzed by citrate synthase in the presence
of L-malate but not D-malate. The ternary complexes with malate reveal the mode
of binding of the substrate acetylCoA in the ground state. The carbonyl oxygen
of the acetyl group is hydrogen bonded to a water molecule and to histidine 274,
allowing unambiguous identification of the orientation of this group. The
structures support the hypothesis that carboxymethyl coenzyme A is a
transition-state analogue for the enolization step of the reaction (Bayer et
al., 1981) and additionally support proposed mechanisms for the condensation
reaction (Karpusas et al., 1990; Alter et al., 1990).

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Pápai,
A.Hamza,
P.M.Pihko,
and
R.K.Wierenga
(2011).
Stereoelectronic requirements for optimal hydrogen-bond-catalyzed enolization.
|
| |
Chemistry,
17,
2859-2866.
|
 |
|
|
|
|
 |
L.C.Kurz,
C.Z.Constantine,
H.Jiang,
and
T.J.Kappock
(2009).
The partial substrate dethiaacetyl-coenzyme A mimics all critical carbon acid reactions in the condensation half-reaction catalyzed by Thermoplasma acidophilum citrate synthase.
|
| |
Biochemistry,
48,
7878-7891.
|
 |
|
|
|
|
 |
D.R.Boutz,
D.Cascio,
J.Whitelegge,
L.J.Perry,
and
T.O.Yeates
(2007).
Discovery of a thermophilic protein complex stabilized by topologically interlinked chains.
|
| |
J Mol Biol,
368,
1332-1344.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.W.van der Kamp,
F.Perruccio,
and
A.J.Mulholland
(2007).
Substrate polarization in enzyme catalysis: QM/MM analysis of the effect of oxaloacetate polarization on acetyl-CoA enolization in citrate synthase.
|
| |
Proteins,
69,
521-535.
|
 |
|
|
|
|
 |
D.M.Anstrom,
K.Kallio,
and
S.J.Remington
(2003).
Structure of the Escherichia coli malate synthase G:pyruvate:acetyl-coenzyme A abortive ternary complex at 1.95 A resolution.
|
| |
Protein Sci,
12,
1822-1832.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.B.Mitchell,
and
J.Smith
(2003).
D-amino acid residues in peptides and proteins.
|
| |
Proteins,
50,
563-571.
|
 |
|
|
|
|
 |
Z.Gu,
D.G.Drueckhammer,
L.Kurz,
K.Liu,
D.P.Martin,
and
A.McDermott
(1999).
Solid state NMR studies of hydrogen bonding in a citrate synthase inhibitor complex.
|
| |
Biochemistry,
38,
8022-8031.
|
 |
|
|
|
|
 |
L.C.Kurz,
T.Nakra,
R.Stein,
W.Plungkhen,
M.Riley,
F.Hsu,
and
G.R.Drysdale
(1998).
Effects of changes in three catalytic residues on the relative stabilities of some of the intermediates and transition states in the citrate synthase reaction.
|
| |
Biochemistry,
37,
9724-9737.
|
 |
|
|
|
|
 |
L.C.Kurz,
J.H.Roble,
T.Nakra,
G.R.Drysdale,
J.M.Buzan,
B.Schwartz,
and
D.G.Drueckhammer
(1997).
Ability of single-site mutants of citrate synthase to catalyze proton transfer from the methyl group of dethiaacetyl-coenzyme A, a non-thioester substrate analog.
|
| |
Biochemistry,
36,
3981-3990.
|
 |
|
|
|
|
 |
C.D.Dickinson,
C.R.Kelly,
and
W.Ruf
(1996).
Identification of surface residues mediating tissue factor binding and catalytic function of the serine protease factor VIIa.
|
| |
Proc Natl Acad Sci U S A,
93,
14379-14384.
|
 |
|
|
|
|
 |
C.S.Poornima,
and
P.M.Dean
(1995).
Hydration in drug design. 3. Conserved water molecules at the ligand-binding sites of homologous proteins.
|
| |
J Comput Aided Mol Des,
9,
521-531.
|
 |
|
|
|
|
 |
P.L.Chau,
and
P.M.Dean
(1994).
Electrostatic complementarity between proteins and ligands. 1. Charge disposition, dielectric and interface effects.
|
| |
J Comput Aided Mol Des,
8,
513-525.
|
 |
|
|
|
|
 |
P.L.Chau,
and
P.M.Dean
(1994).
Electrostatic complementarity between proteins and ligands. 2. Ligand moieties.
|
| |
J Comput Aided Mol Des,
8,
527-544.
|
 |
|
|
|
|
 |
A.J.Patton,
D.W.Hough,
P.Towner,
and
M.J.Danson
(1993).
Does Escherichia coli possess a second citrate synthase gene?
|
| |
Eur J Biochem,
214,
75-81.
|
 |
|
|
|
|
 |
S.L.Moodie,
and
J.M.Thornton
(1993).
A study into the effects of protein binding on nucleotide conformation.
|
| |
Nucleic Acids Res,
21,
1369-1380.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

