Protein-tyrosine-phosphatase non-receptor class
The protein tyrosine phosphatases are a group of enzymes that remove the phosphate groups from tyrosine residues. These enzymes are important in cell signalling during growth and development and act antagonistically with the protein tyrosine kinases.
The PTP family is subdivided into several groups including the tyrosine specific receptor and non-receptor-like enzymes, the dual-specificity group, the low molecular weight PTPs and the cdc25 group. All members share the same mechanism of hydrolysis and are characterised by a CX5R sequence motif. The CX5R motif forms a loop that provides an oxyanion hole for the stabilisation of the phosphate group. Binding to this loop causes the movement of a conserved aspartic acid into the active site. However the cdc25 group is highly divergent. A cysteine-phosphate intermediate is formed during the reaction and is then hydrolysed. Although the different groups of PTPs are relatively divergent their active sites are highly conserved.
The Yersinia genus contains a plasmid encoding a virulence factor, YopH, a tyrosine phosphatase which translocates itself into the host cell using the N-terminus. YopH is hyperactive compared to human PTPases. The catalytic domains are structurally homologous to human PTP1B, which is also a tyrosine phosphatase. YopH cleaves the phosphate off phosphorylated tyrosine residues on host proteins, interrupting the control of many eukaryotic cellular processes, and contributes to the ability to resist phagocytosis by peritoneal macrophages.
Reference Protein and Structure
- Sequence
-
P15273
 (3.1.3.48)
(3.1.3.48)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Yersinia enterocolitica (Bacteria)

- PDB
-
1ytw
- YERSINIA PTPASE COMPLEXED WITH TUNGSTATE
(2.4 Å)



- Catalytic CATH Domains
-
3.90.190.10
 (see all for 1ytw)
(see all for 1ytw)
- Cofactors
- Water (1)
Enzyme Reaction (EC:3.1.3.48)
Enzyme Mechanism
Introduction
The mechanism is thought to be primarily dissociative, with more catalytic importance placed on stabilising the negative charge on the leaving group, rather than activation of the nucleophile. Phosphoryl transfer occurs in a single step, with an inline displacement mechanism. The key components are as follows:
- Thr 410 hydrogen bonds to Cys 403, lowering Cys 403's pKa and stabilising a thiolate anion on this residue.
- Asp 356 is protonated, and acts as a general acid to protonate the substrate tyrosine residue, promoting phosphoryl transfer.
- The trigonal bipyramid transition state is stabilised by Arg 409.
- Cys 403 becomes phosphorylated. The substrate is released and water enters the active site.
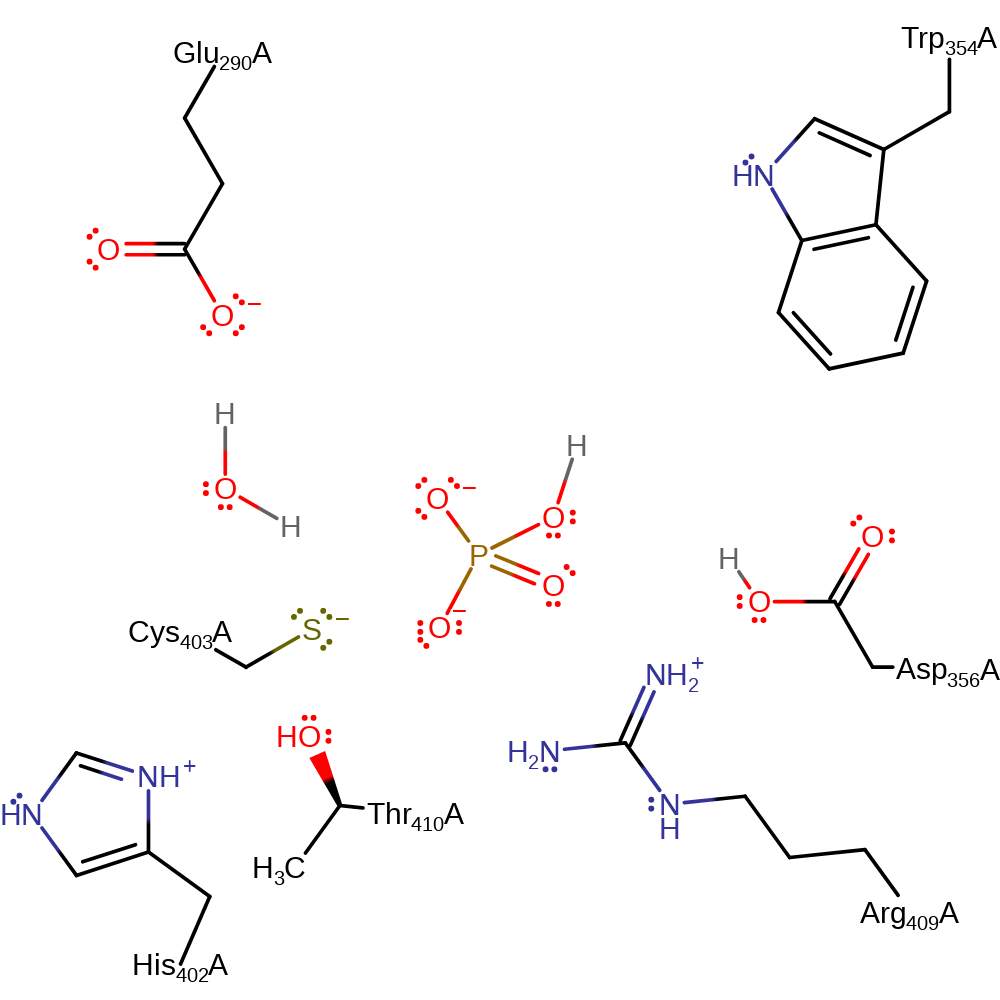
- Asp 356 activates the water molecule by hydrogen bonding.
- During phosphoryl transfer, Thr 410 stabilises the growing negative charge on the thiolate of Cys 403. Arg 409 again stabilises the transition state. Asp 356 deprotonates the water nucleophile.
- Inorganic phosphate is formed.
Catalytic Residues Roles
| UniProt | PDB* (1ytw) | ||
| His402 | His402(240)A | Stabilises the negatively charged intermediates and transition states. | hydrogen bond donor, electrostatic stabiliser |
| Asp356 | Asp356(194)A | Acts as an general acid-base catalyst, protonating the substrate and deprotonating the nucleophilic water molecule. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Cys403 | Cys403(241)A | Nucleophile that removes the phosphoryl group from the substrate. | nucleofuge, nucleophile |
| Arg409 | Arg409(247)A | Stabilises the transition states, through coplanar, divalent guanidium-phosphate interactions. | hydrogen bond donor, electrostatic stabiliser |
| Thr410 | Thr410(248)A | Lowers the pKa of the nucleophilic Cys, and stabilises the thiolate anion as a leaving group during phosphoenzyme hydrolysis | hydrogen bond donor, electrostatic stabiliser |
| Glu290 | Glu290(128)A | Helps position the substrate through an active site water molecule. | steric role |
| Trp354 | Trp354(192)A | Although not a catalytic residue in the traditional sense, this residue is critical in the loop movement upon substrate binding which places the general acid/base Asp356 for catalysis. Mutation of this residue severely impairs catalysis. | steric role |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, enzyme-substrate complex formation, intermediate formation, proton transfer, enzyme-substrate complex cleavage, dephosphorylation, intermediate collapse, intermediate terminated, hydrolysisReferences
- Hoff RH et al. (1999), J Am Chem Soc, 121, 9514-9521. Does Positive Charge at the Active Sites of Phosphatases Cause a Change in Mechanism? The Effect of the Conserved Arginine on the Transition State for Phosphoryl Transfer in the Protein-Tyrosine Phosphatase fromYersinia. DOI:10.1021/ja992361o.
- Moise G et al. (2015), Biochemistry, 54, 6490-6500. Conservative Tryptophan Mutants of the Protein Tyrosine Phosphatase YopH Exhibit Impaired WPD-Loop Function and Crystallize with Divanadate Esters in Their Active Sites. DOI:10.1021/acs.biochem.5b00496. PMID:26445170.
- Ke S et al. (2012), J Phys Chem B, 116, 6166-6176. Investigation of Catalytic Loop Structure, Dynamics, and Function Relationship ofYersiniaProtein Tyrosine Phosphatase by Temperature-Jump Relaxation Spectroscopy and X-ray Structural Determination. DOI:10.1021/jp3037846. PMID:22564106.
- Brandão TA et al. (2009), J Am Chem Soc, 131, 778-786. Impaired Acid Catalysis by Mutation of a Protein Loop Hinge Residue in a YopH Mutant Revealed by Crystal Structures. DOI:10.1021/ja807418b. PMID:19140798.
- Zhang ZY (2003), Prog Nucleic Acid Res Mol Biol, 73, 171-220. Mechanistic Studies on Protein Tyrosine Phosphatases. DOI:10.1016/s0079-6603(03)01006-7. PMID:12882518.
- Wang F et al. (1998), Biochemistry, 37, 15289-15299. Conformational and Dynamic Changes ofYersiniaProtein Tyrosine Phosphatase Induced by Ligand Binding and Active Site Mutation and Revealed by H/D Exchange and Electrospray Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry†. DOI:10.1021/bi981481q. PMID:9799489.
- Pannifer AD et al. (1998), J Biol Chem, 273, 10454-10462. Visualization of the Cysteinyl-phosphate Intermediate of a Protein-tyrosine Phosphatase by X-ray Crystallography. DOI:10.1074/jbc.273.17.10454. PMID:9553104.
- Fauman EB et al. (1996), J Biol Chem, 271, 18780-18788. The X-ray Crystal Structures of Yersinia Tyrosine Phosphatase with Bound Tungstate and Nitrate: MECHANISTIC IMPLICATIONS. DOI:10.1074/jbc.271.31.18780. PMID:8702535.
- Fauman EB et al. (1996), Trends Biochem Sci, 21, 413-417. Structure and function of theprotein tyrosine phosphatases. DOI:10.1016/s0968-0004(96)10059-1.
- Fauman EB et al. (1996), Trends Biochem Sci, 21, 185-200. Structure and function of the T-cell protein tyrosine phosphatase. DOI:10.1007/978-3-540-40035-6_10. PMID:8987394.
- Zhang ZY et al. (1995), Biochemistry, 34, 16389-16396. Catalytic function of the conserved hydroxyl group in the protein tyrosine phosphatase signature motif. DOI:10.1021/bi00050a020. PMID:8845365.
- Zhang ZY et al. (1994), Proc Natl Acad Sci U S A, 91, 1624-1627. Dissecting the catalytic mechanism of protein-tyrosine phosphatases. DOI:10.1073/pnas.91.5.1624. PMID:8127855.
- Zhang ZY et al. (1994), Biochemistry, 33, 15266-15270. The Cys(X)5Arg Catalytic Motif in Phosphoester Hydrolysis. DOI:10.1021/bi00255a007. PMID:7803389.
- Stuckey JA et al. (1994), Nature, 370, 571-575. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 Å and the complex with tungstate. DOI:10.1038/370571a0. PMID:8052312.
- Guan KL et al. (1991), J Biol Chem, 266, 17026-17030. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. PMID:1654322.
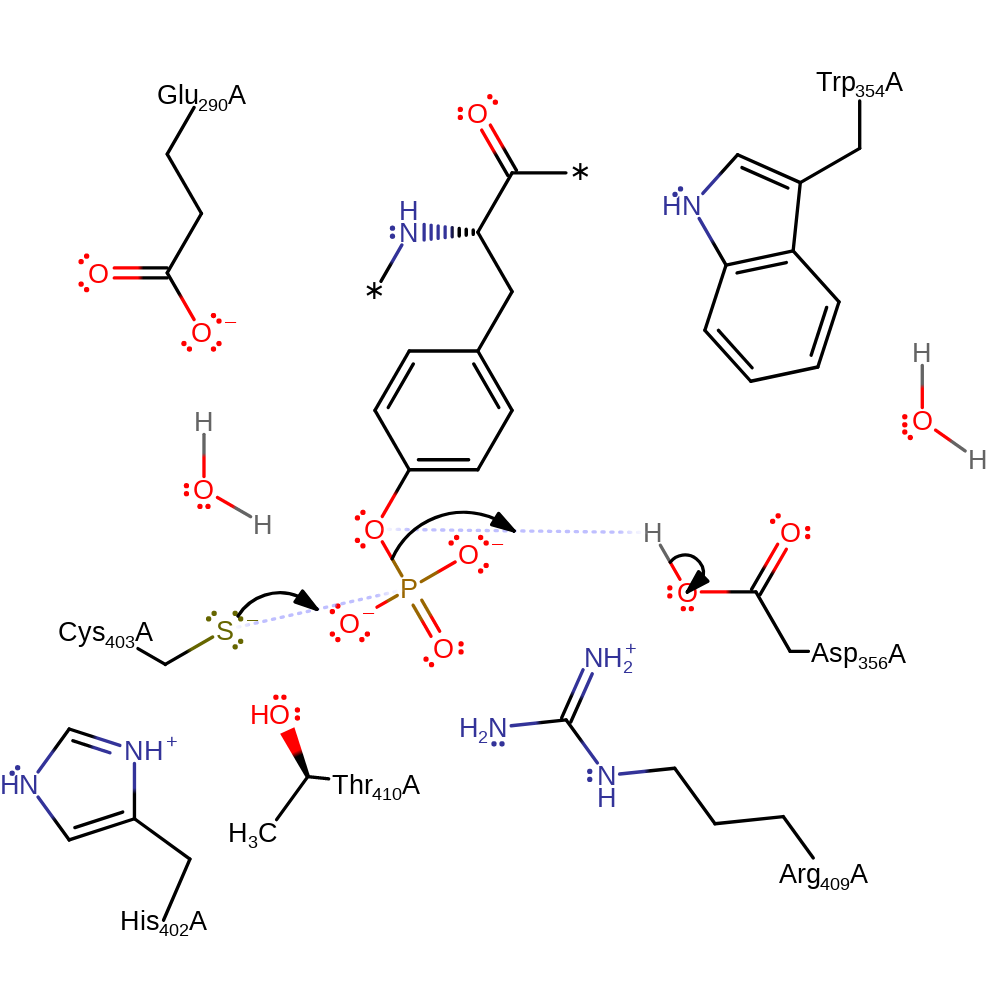
Step 1. Cys403 initiates a nucleophilic attack upon the phosphate of the substrate in a substitution reaction, which eliminates the tyrosine with concomitant deprotonation of Asp356.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr410(248)A | electrostatic stabiliser, hydrogen bond donor |
| His402(240)A | electrostatic stabiliser, hydrogen bond donor |
| Arg409(247)A | hydrogen bond donor, electrostatic stabiliser |
| Asp356(194)A | hydrogen bond donor |
| Glu290(128)A | steric role |
| Trp354(192)A | steric role |
| Asp356(194)A | proton donor |
| Cys403(241)A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, enzyme-substrate complex formation, intermediate formation, proton transfer
Step 2. Asp356 deprotonates water, which initiates a nucleophilic attack upon the phosphate of the covalently bound intermediate in a substitution reaction, which eliminates Cys403.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg409(247)A | hydrogen bond donor, electrostatic stabiliser |
| His402(240)A | electrostatic stabiliser, hydrogen bond donor |
| Thr410(248)A | hydrogen bond donor, electrostatic stabiliser |
| Asp356(194)A | hydrogen bond acceptor |
| Glu290(128)A | steric role |
| Trp354(192)A | steric role |
| Cys403(241)A | nucleofuge |
| Asp356(194)A | proton acceptor |




 Download:
Download: