Beta-lactamase (Class A)
The Class A Beta lactamase acts to hydrolyse the endocyclic amide bond of beta-lactam substrates in a two step mechanism: acylation (covalent attachment of the beta-lactam to an active site serine, Ser70), followed by deacylation. The TEM-type are the most prevalent beta-lactamases in enterobacteria and confer resistance to penicillins and cephalosporins in these species.
TEM-3 and TEM-4 are capable of hydrolyzing cefotaxime and ceftazidime. TEM-5 is capable of hydrolysing ceftazidime. TEM-6 is capable of hydrolysing ceftazidime and aztreonam. TEM-8/CAZ-2, TEM-16/CAZ-7 and TEM-24/CAZ-6 are markedly active against ceftazidime. IRT-4 shows resistance to beta-lactamase inhibitors.
Reference Protein and Structure
- Sequence
-
P62593
 (3.5.2.6)
(3.5.2.6)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli (Bacteria)

- PDB
-
1btl
- CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION
(1.8 Å)



- Catalytic CATH Domains
-
3.40.710.10
 (see all for 1btl)
(see all for 1btl)
- Cofactors
- Water (1)
Enzyme Reaction (EC:3.5.2.6)
Enzyme Mechanism
Introduction
The beta-lactamase mechanism consists of two steps: acylation (covalent attachment of the beta-lactam to an active site serine, Ser70), followed by deacylation. The first step, base catalysed nucleophilic attack from Ser70 at the carbonyl carbon of the lactam occurs where activation of Ser70 by Glu166 and nucleophilic attack happen simultaneously. The protonation at lactam N(1) is catalysed within a hydrogen bonded cluster involving the 2-carboxylate group in the substrate, side chains Ser130, Lys234 and a exogenous solvent molecule. The nucleophilic Ser70 has been shown to approach the butterfly cadge beta-lactam structure from the exo face, its activity directed by interactions with the surrounding ion pairs.
Catalytic Residues Roles
| UniProt | PDB* (1btl) | ||
| Ser68 | Ser70(45)A | The residue side chain acts as a nucleophile towards the beta lactam substrate carbonyl group, with activation from acid/base interaction with a conserved hydrolytic water molecule, in turn interacting with Glu166. The backbone amide acts as part of an oxyanion hole, stabilising the transition state. The residue forms an ion pair with Lys 73. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleophile, proton acceptor, proton donor, nucleofuge, electrostatic stabiliser |
| Lys71 | Lys73(48)A | The residue forms an ion pair with Ser 70,which is involved in the proton transfer event.The residue remains protonated through out the reaction, and is thought to be involved in directing the Ser 70 hydroxyl group for effective catalysis. The residue interacts with Glu 166 via hydrogen bonding. | hydrogen bond acceptor, hydrogen bond donor |
| Ser128 | Ser130(105)A | The residue is implicated in catalytic action within a hydrogen bonding network which mediates protonation of the substrate nitrogen through proton relay.The residue interacts with K234 through hydrogen bonding and the substrate oxygen 12, held within the carboxylate group. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator, electrostatic stabiliser |
| Glu164 | Glu166(141)A | In acylation the residue acts as a general base towards a structurally conserved water molecule,leading to the deprotonation of Ser70 (proton relay). In deacylation, Glu 166 abstracts a proton from a water molecule, activating a nucleophile for attack at the substrate carbon linked to the gamma oxygen of S 70. The residue is hydrogen bonded to Lys 73. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator, electrostatic stabiliser |
| Lys232 | Lys234(209)A | The residue is implicated in the initial protonation of the substrate N(1) by interaction with S130 through hydrogen bonding. | electrostatic stabiliser |
| Ala235 (main-N), Ser68 (main-N) | Ala237(212)A (main-N), Ser70(45)A (main-N) | Backbone of the residue forms an oxyanion hole in conjunction with the backbone of residue S70 to stabilise the anionic tetrahedral intermediate. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, overall product formed, enzyme-substrate complex cleavage, native state of enzyme regenerated, intermediate collapse, intermediate terminatedReferences
- Chen Y et al. (2007), J Am Chem Soc, 129, 5378-5380. The Acylation Mechanism of CTX-M β-Lactamase at 0.88 Å Resolution. DOI:10.1021/ja0712064. PMID:17408273.
- Hermann JC et al. (2006), Org Biomol Chem, 4, 206-210. Molecular mechanisms of antibiotic resistance: QM/MM modelling of deacylation in a class A β-lactamase. DOI:10.1039/b512969a. PMID:16391762.
- Hermann JC et al. (2003), J Am Chem Soc, 125, 9590-9591. Identification of Glu166 as the General Base in the Acylation Reaction of Class A β-Lactamases through QM/MM Modeling. DOI:10.1021/ja034434g. PMID:12904016.
- Nukaga M et al. (2003), J Mol Biol, 328, 289-301. Ultrahigh Resolution Structure of a Class A β-Lactamase: On the Mechanism and Specificity of the Extended-spectrum SHV-2 Enzyme. DOI:10.1016/s0022-2836(03)00210-9.
- Castillo R et al. (2002), J Am Chem Soc, 124, 1809-1816. Role of Protein Flexibility in Enzymatic Catalysis: Quantum Mechanical−Molecular Mechanical Study of the Deacylation Reaction in Class A β-Lactamases. DOI:10.1021/ja017156z. PMID:11853460.
- Atanasov BP et al. (2000), Proc Natl Acad Sci U S A, 97, 3160-3165. Protonation of the beta -lactam nitrogen is the trigger event in the catalytic action of class A beta -lactamases. DOI:10.1073/pnas.060027897. PMID:10716727.
- MATAGNE A et al. (1998), Biochem J, 330, 581-598. Catalytic properties of class A β-lactamases: efficiency and diversity. DOI:10.1042/bj3300581.
- Guillaume G et al. (1997), J Biol Chem, 272, 5438-5444. Site-directed Mutagenesis of Glutamate 166 in Two -Lactamases: KINETIC AND MOLECULAR MODELING STUDIES. DOI:10.1074/jbc.272.9.5438.
- Chen CC et al. (1996), Biochemistry, 35, 12251-12258. Structure and Kinetics of the β-Lactamase Mutants S70A and K73H fromStaphylococcus aureusPC1†,‡. DOI:10.1021/bi961153v. PMID:8823158.
- Damblon C et al. (1996), Proc Natl Acad Sci U S A, 93, 1747-1752. The catalytic mechanism of beta-lactamases: NMR titration of an active-site lysine residue of the TEM-1 enzyme. DOI:10.1073/pnas.93.5.1747. PMID:8700829.
- Strynadka NC et al. (1992), Nature, 359, 700-705. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. DOI:10.1038/359700a0. PMID:1436034.
- Lamotte-Brasseur J et al. (1991), Biochem J, 279, 213-221. Mechanism of acyl transfer by the class A serineβ-lactamase ofStreptomyces albusG. DOI:10.1042/bj2790213. PMID:1930139.
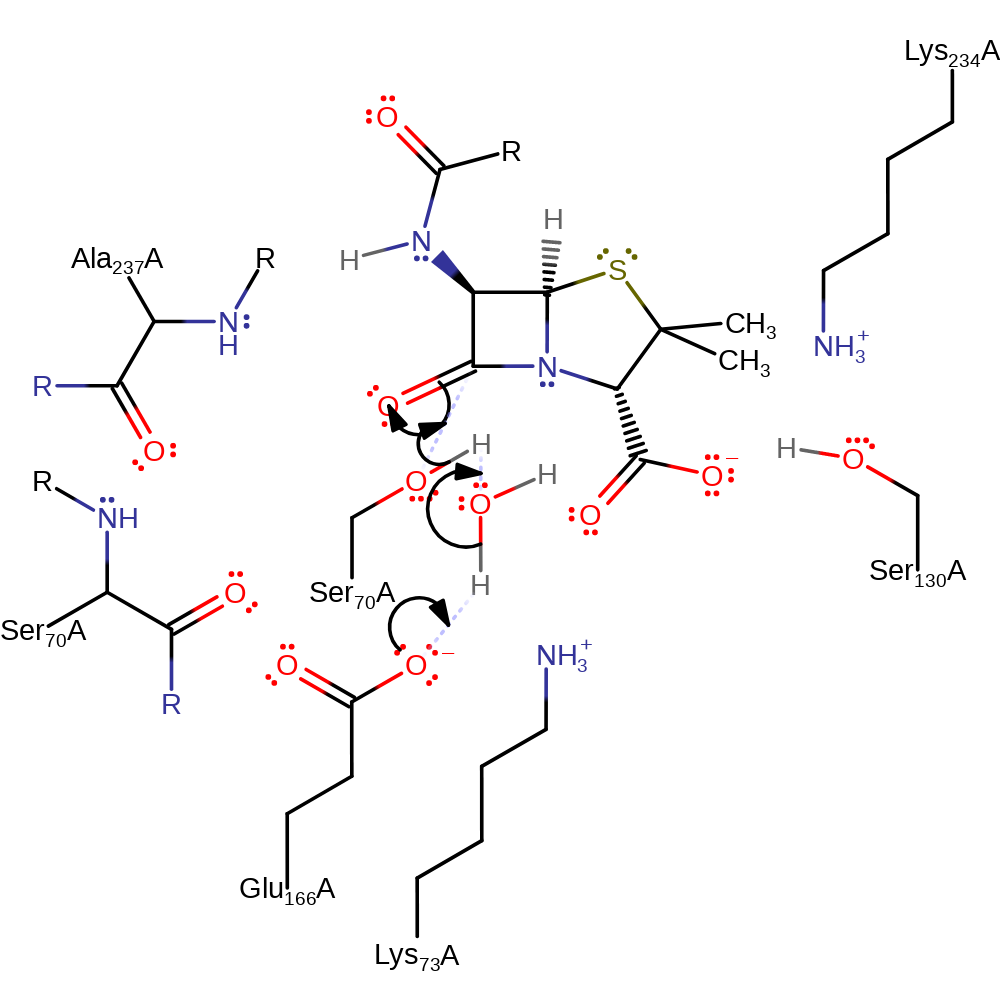
Step 1. Glu166 deprotonates a conserved water molecule which in turn deprotonates the nucleophilic Ser70, initiating the nucleophilic addition onto the carbonyl carbon of the beta-lactam, forming a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys73(48)A | hydrogen bond acceptor |
| Ser130(105)A | hydrogen bond donor, hydrogen bond acceptor, activator |
| Ser70(45)A | hydrogen bond donor |
| Glu166(141)A | hydrogen bond acceptor, activator |
| Ser70(45)A | electrostatic stabiliser |
| Ala237(212)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Lys234(209)A | electrostatic stabiliser |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Ser70(45)A | proton donor |
| Glu166(141)A | proton acceptor |
| Ser70(45)A | nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used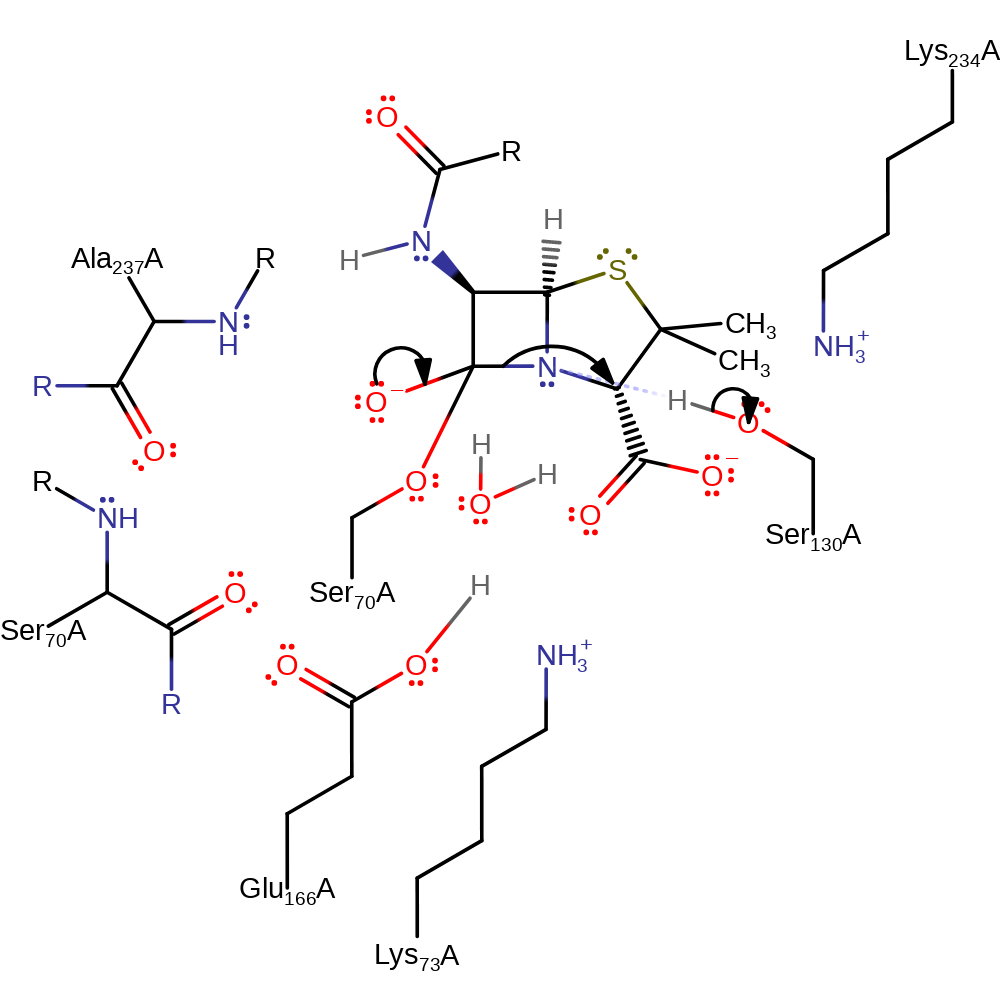
Step 2. The tetrahedral intermediate collapses, cleaving the C-N bond in the beta-lactam, the nitrogen deprotonates Ser130.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys234(209)A | electrostatic stabiliser |
| Lys73(48)A | hydrogen bond donor |
| Ser130(105)A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Ser70(45)A | covalently attached, hydrogen bond acceptor |
| Glu166(141)A | hydrogen bond acceptor, electrostatic stabiliser |
| Ser70(45)A | hydrogen bond donor, electrostatic stabiliser |
| Ala237(212)A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Ser130(105)A | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys234(209)A | electrostatic stabiliser |
| Lys73(48)A | hydrogen bond donor |
| Ser130(105)A | hydrogen bond acceptor |
| Ser70(45)A | covalently attached, hydrogen bond acceptor |
| Glu166(141)A | hydrogen bond acceptor |
| Ser70(45)A | hydrogen bond donor |
| Ala237(212)A (main-N) | hydrogen bond donor |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Ser130(105)A | proton acceptor |
| Glu166(141)A | proton donor |
Chemical Components
proton transfer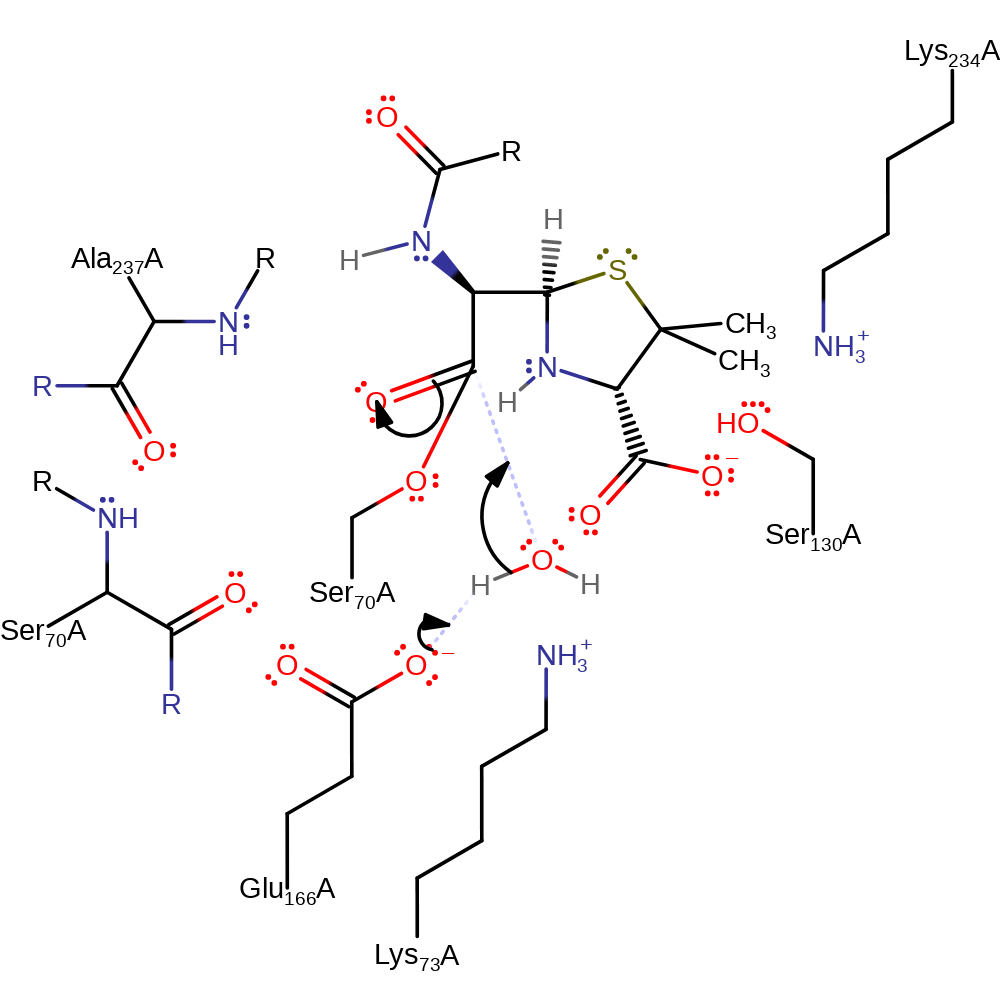
Step 4. Glu166 deprotonates water, which initiates a nucleophilic addition at the carbonyl carbon, forming a new tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys73(48)A | hydrogen bond donor |
| Ser130(105)A | hydrogen bond donor, hydrogen bond acceptor |
| Ser70(45)A | covalently attached |
| Glu166(141)A | hydrogen bond acceptor |
| Ala237(212)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser70(45)A | hydrogen bond donor, electrostatic stabiliser |
| Lys234(209)A | electrostatic stabiliser |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Glu166(141)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation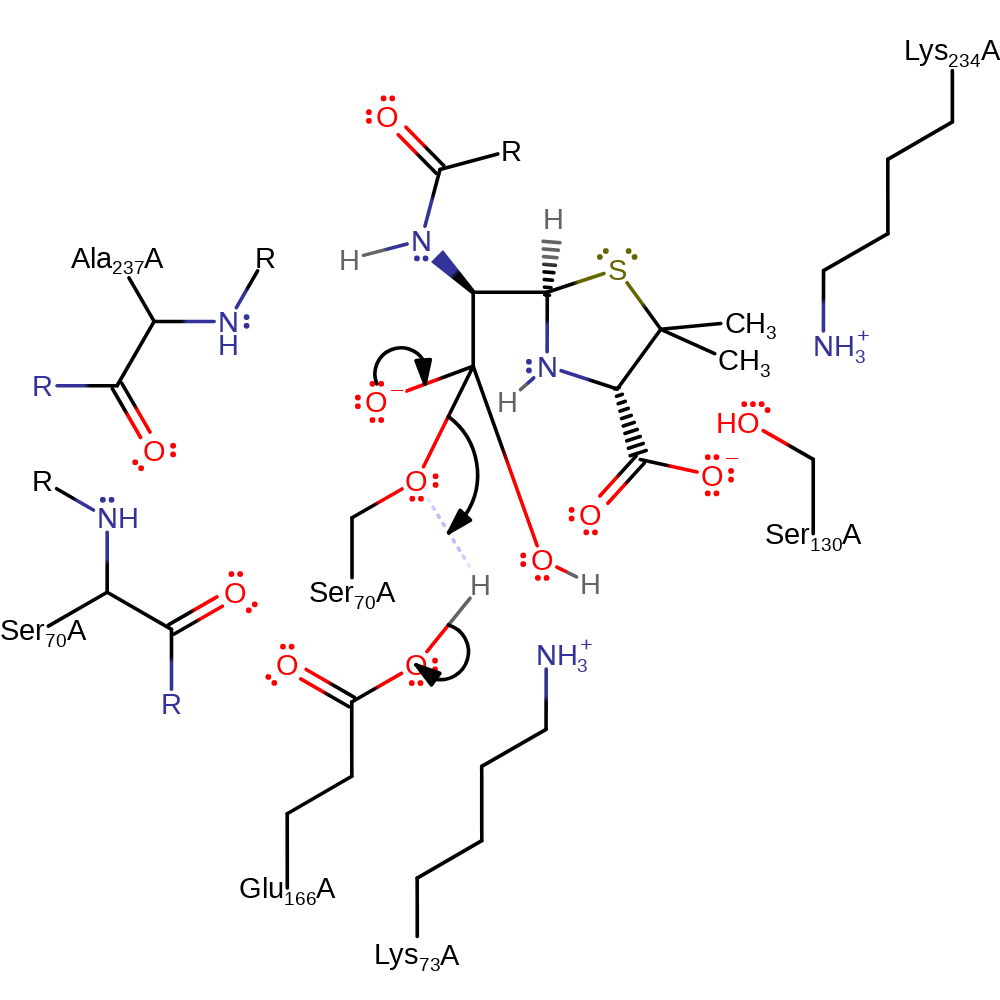
Step 5. The tetrahedral intermediate collapses, cleaving the acyl-enzyme bond and liberating Ser70, which in turn deprotonates the Glu166.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys73(48)A | hydrogen bond donor |
| Ser130(105)A | hydrogen bond donor, hydrogen bond acceptor |
| Ser70(45)A | hydrogen bond acceptor |
| Glu166(141)A | hydrogen bond donor, hydrogen bond acceptor |
| Ser70(45)A | hydrogen bond donor, electrostatic stabiliser |
| Ala237(212)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Lys234(209)A | electrostatic stabiliser |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Ser70(45)A | proton acceptor, nucleofuge |
| Glu166(141)A | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, overall product formed, enzyme-substrate complex cleavage, native state of enzyme regenerated, intermediate collapse, intermediate terminatedIntroduction
The beta-lactamase mechanism consists of two steps: acylation (covalent attachment of the beta-lactam to an active site serine, Ser70), followed by deacylation. In the first step, a base catalysed nucleophilic attack from Ser70 at the carbonyl carbon of the beta-lactam occurs, where activation of Ser70 by Lys73 and nucleophilic attack happen simultaneously. The protonation at lactam N(1) is catalysed within a hydrogen bonded cluster involving the 2-carboxylate group in the substrate, side chains Ser130, Lys234 and a exogenous solvent molecule. The nucleophilic Ser70 has been shown to approach the butterfly cadge beta-lactam structure from the exo face, its activity directed by interactions with the surrounding ion pairs.
Catalytic Residues Roles
| UniProt | PDB* (1btl) | ||
| Ser68 | Ser70(45)A | Main chain amide forms part of the oxyanion hole. Side chain acts as the catalytic nucleophile. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleophile, proton acceptor, proton donor, nucleofuge, electrostatic stabiliser |
| Lys71 | Lys73(48)A | Acts as a general acid/base. Deprotonates the Ser70 side chain, activating it for acylation. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Ser128 | Ser130(105)A | Acts as a general acid/base. Furnishes the substrate N with a proton, and reprotonates from Lys73. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator, electrostatic stabiliser |
| Glu164 | Glu166(141)A | Acts as a general acid/base, deprotonating water for the deacylation step. It is deprotonated by Ser70 in the final step of the reaction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator, electrostatic stabiliser |
| Lys232 | Lys234(209)A | No explicit role in this mechanism proposal. Likely involved in stabilising the substrate. | electrostatic stabiliser |
| Ala235 (main-N), Ser68 (main-N) | Ala237(212)A (main-N), Ser70(45)A (main-N) | Backbone of the residue forms an oxyanion hole to stabilise the anionic tetrahedral intermediate. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, proton transfer, unimolecular elimination by the conjugate base, overall product formed, enzyme-substrate complex cleavage, native state of enzyme regenerated, intermediate collapse, intermediate terminatedReferences
- Maveyraud L et al. (2000), Structure, 8, 1289-1298. Insights into Class D β-Lactamases Are Revealed by the Crystal Structure of the OXA10 Enzyme from Pseudomonas aeruginosa. DOI:10.1016/s0969-2126(00)00534-7. PMID:11188693.
- Chudyk EI et al. (2014), Chem Commun (Camb), 50, 14736-14739. QM/MM simulations as an assay for carbapenemase activity in class A β-lactamases. DOI:10.1039/C4CC06495J. PMID:25321894.
- Hermann JC et al. (2003), J Am Chem Soc, 125, 9590-9591. Identification of Glu166 as the General Base in the Acylation Reaction of Class A β-Lactamases through QM/MM Modeling. DOI:10.1021/ja034434g. PMID:12904016.
- Castillo R et al. (2002), J Am Chem Soc, 124, 1809-1816. Role of Protein Flexibility in Enzymatic Catalysis: Quantum Mechanical−Molecular Mechanical Study of the Deacylation Reaction in Class A β-Lactamases. DOI:10.1021/ja017156z. PMID:11853460.
- Atanasov BP et al. (2000), Proc Natl Acad Sci U S A, 97, 3160-3165. Protonation of the beta -lactam nitrogen is the trigger event in the catalytic action of class A beta -lactamases. DOI:10.1073/pnas.060027897. PMID:10716727.
- Pitarch J et al. (2000), Journal of the Chemical Society, Perkin Transactions 2, 761-767. A quantum mechanics/molecular mechanics study of the acylation reaction of TEM1 β-lactamase and penicillanate. DOI:10.1039/a908264f.
- Maveyraud L et al. (1998), Biochemistry, 37, 2622-2628. Crystal Structure of an Acylation Transition-State Analog of the TEM-1 β-Lactamase. Mechanistic Implications for Class A β-Lactamases†. DOI:10.1021/bi972501b. PMID:9485412.
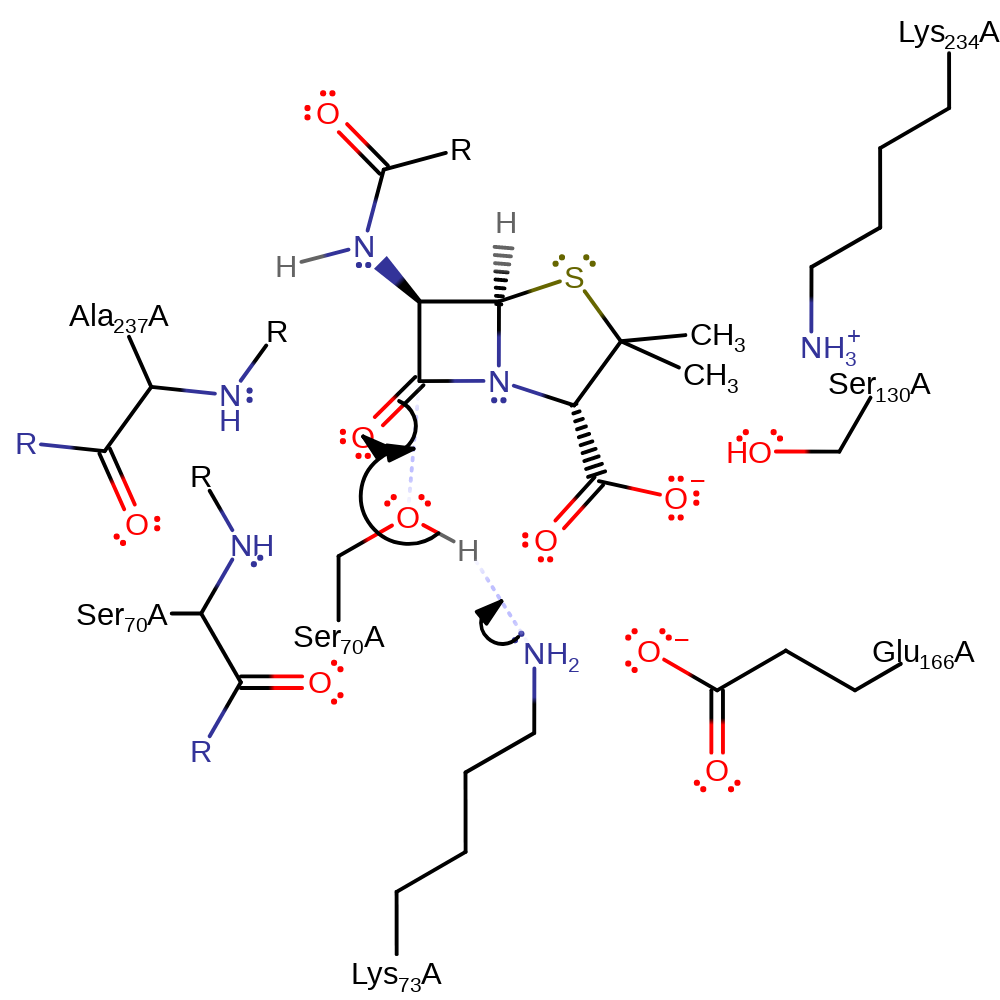
Step 1. Lys73 deprotonates Ser70, which initiates a nucleophilic addition onto the carbonyl carbon of the beta-lactam, forming a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys73(48)A | hydrogen bond acceptor |
| Ser130(105)A | hydrogen bond donor, hydrogen bond acceptor, activator |
| Ser70(45)A | hydrogen bond donor |
| Glu166(141)A | hydrogen bond acceptor, activator |
| Ser70(45)A | electrostatic stabiliser |
| Ala237(212)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Lys234(209)A | electrostatic stabiliser |
| Lys73(48)A | proton acceptor |
| Ser70(45)A | proton donor, nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, proton transfer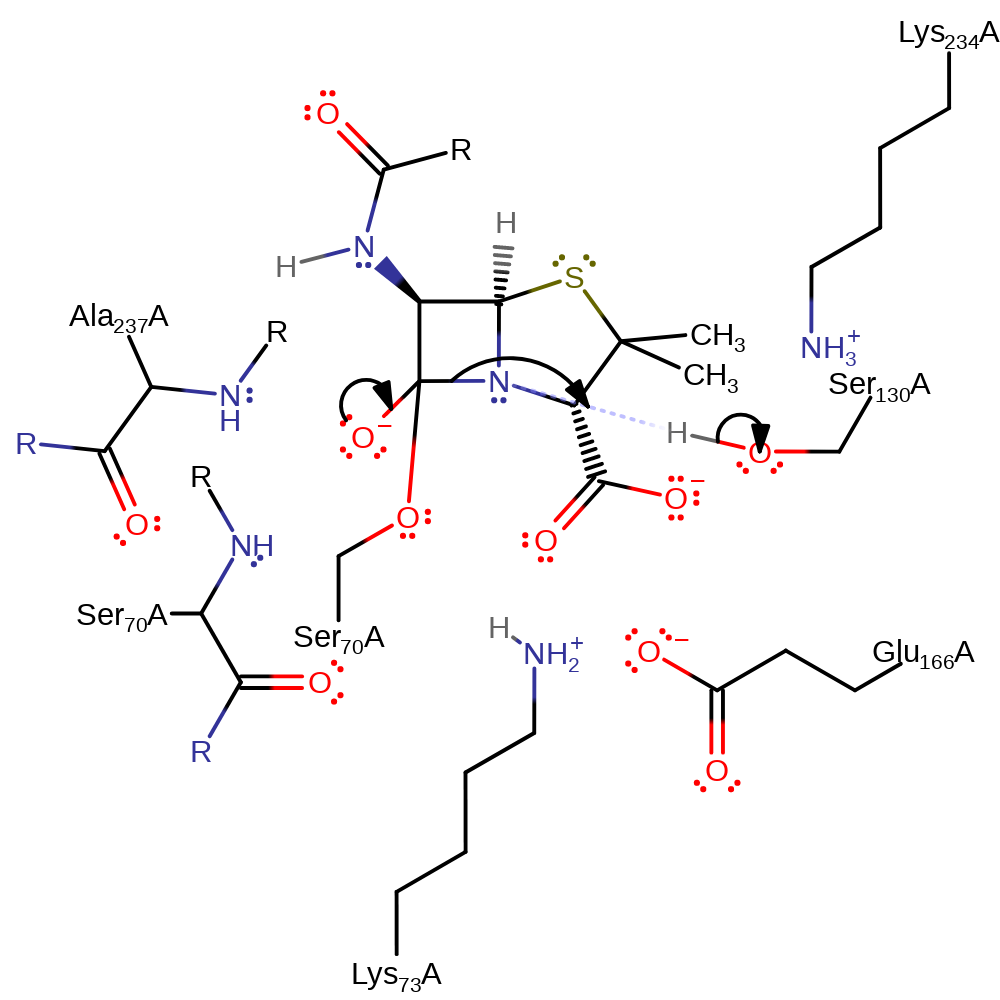
Step 2. The tetrahedral intermediate collapses, cleaving the C-N bond in the beta-lactam, the nitrogen deprotonates Ser130.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys73(48)A | hydrogen bond donor |
| Ser130(105)A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Ser70(45)A | covalently attached, hydrogen bond acceptor |
| Glu166(141)A | hydrogen bond acceptor, electrostatic stabiliser |
| Ser70(45)A | hydrogen bond donor, electrostatic stabiliser |
| Ala237(212)A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Lys234(209)A | electrostatic stabiliser |
| Ser130(105)A | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys73(48)A | hydrogen bond donor |
| Ser130(105)A | hydrogen bond acceptor |
| Ser70(45)A | covalently attached, hydrogen bond acceptor |
| Glu166(141)A | hydrogen bond acceptor |
| Ser70(45)A | hydrogen bond donor |
| Ala237(212)A (main-N) | hydrogen bond donor |
| Lys234(209)A | electrostatic stabiliser |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Ser130(105)A | proton acceptor |
| Lys73(48)A | proton donor |
Chemical Components
proton transfer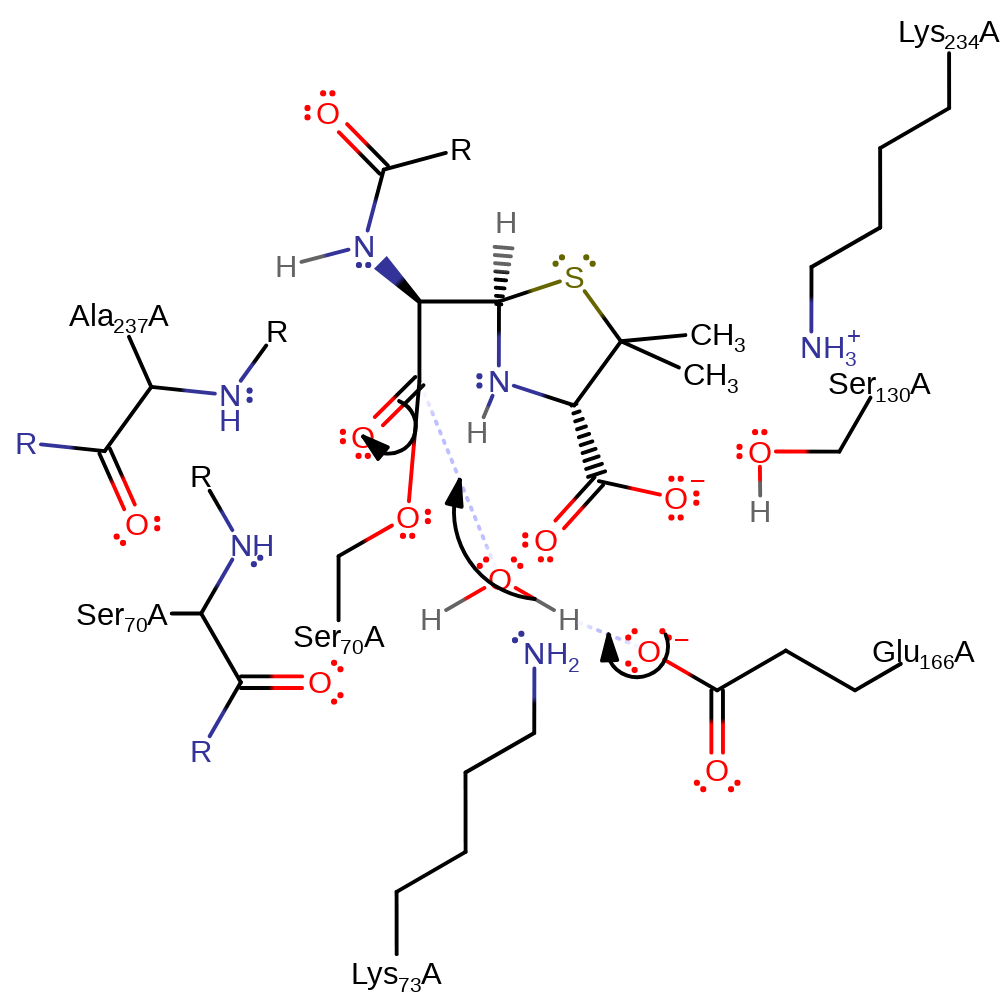
Step 4. Glu166 deprotonates water, which initiates a nucleophilic addition at the carbonyl carbon, forming a new tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys73(48)A | hydrogen bond donor |
| Ser130(105)A | hydrogen bond donor, hydrogen bond acceptor |
| Ser70(45)A | covalently attached |
| Glu166(141)A | hydrogen bond acceptor |
| Ala237(212)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser70(45)A | hydrogen bond donor, electrostatic stabiliser |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Lys234(209)A | electrostatic stabiliser |
| Glu166(141)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation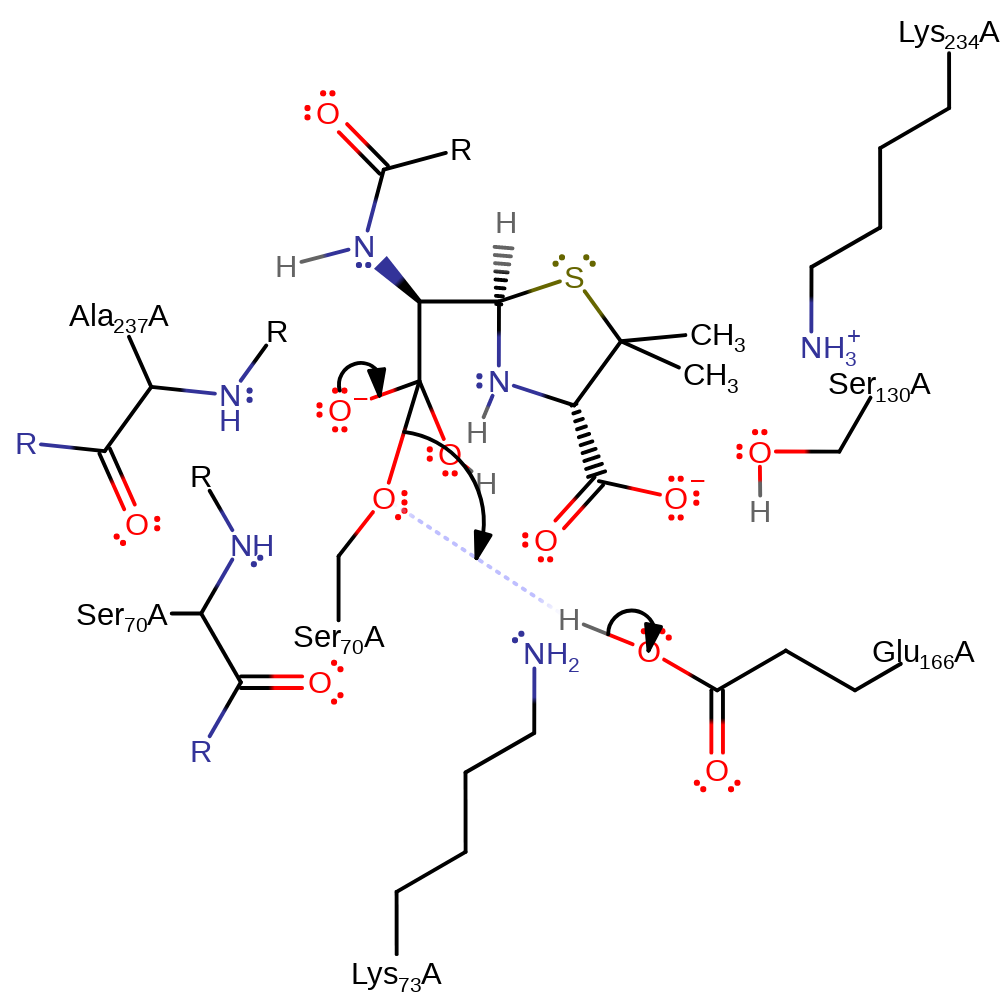
Step 5. The tetrahedral intermediate collapses, cleaving the acyl-enzyme bond and liberating Ser70, which in turn deprotonates the Glu166.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys73(48)A | hydrogen bond donor |
| Ser130(105)A | hydrogen bond donor, hydrogen bond acceptor |
| Ser70(45)A | hydrogen bond acceptor |
| Glu166(141)A | hydrogen bond donor, hydrogen bond acceptor |
| Ser70(45)A | hydrogen bond donor, electrostatic stabiliser |
| Ala237(212)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Ser70(45)A (main-N) | electrostatic stabiliser |
| Lys234(209)A | electrostatic stabiliser |
| Ser70(45)A | proton acceptor, nucleofuge |
| Glu166(141)A | proton donor |



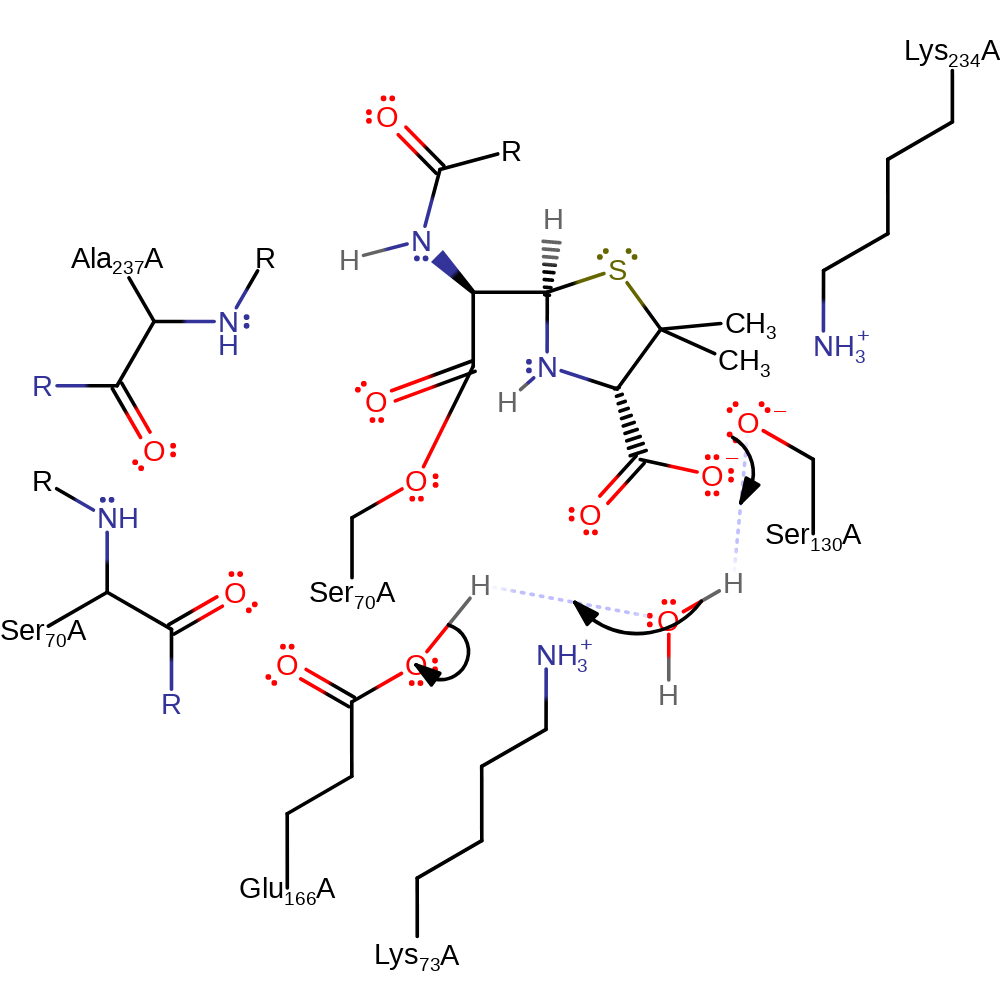
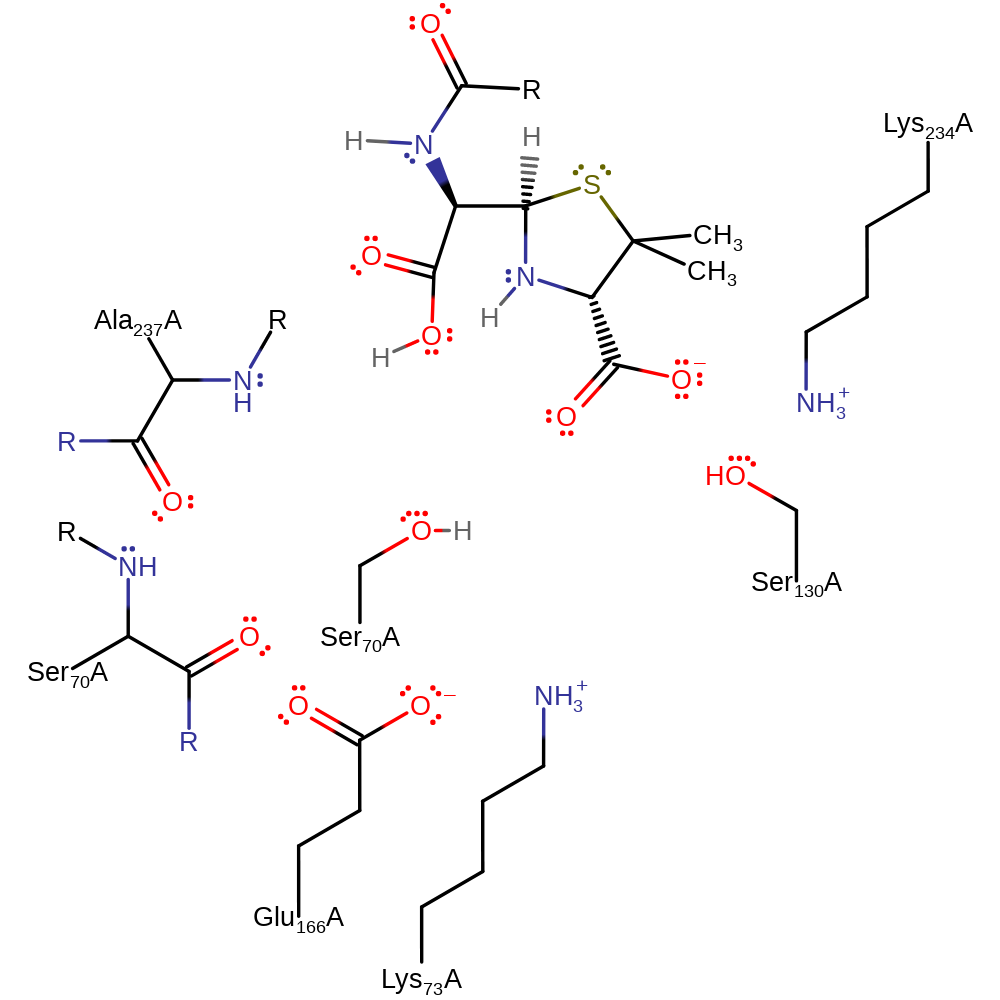 Download:
Download: 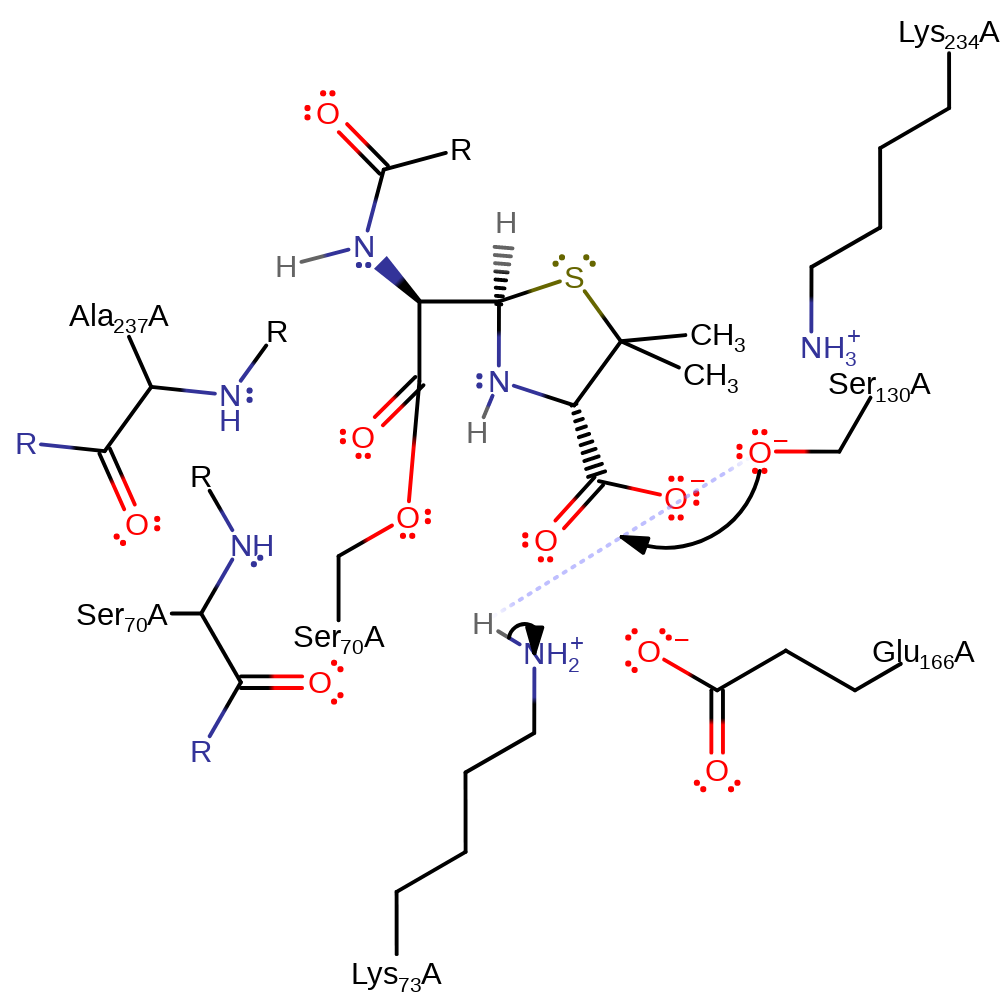
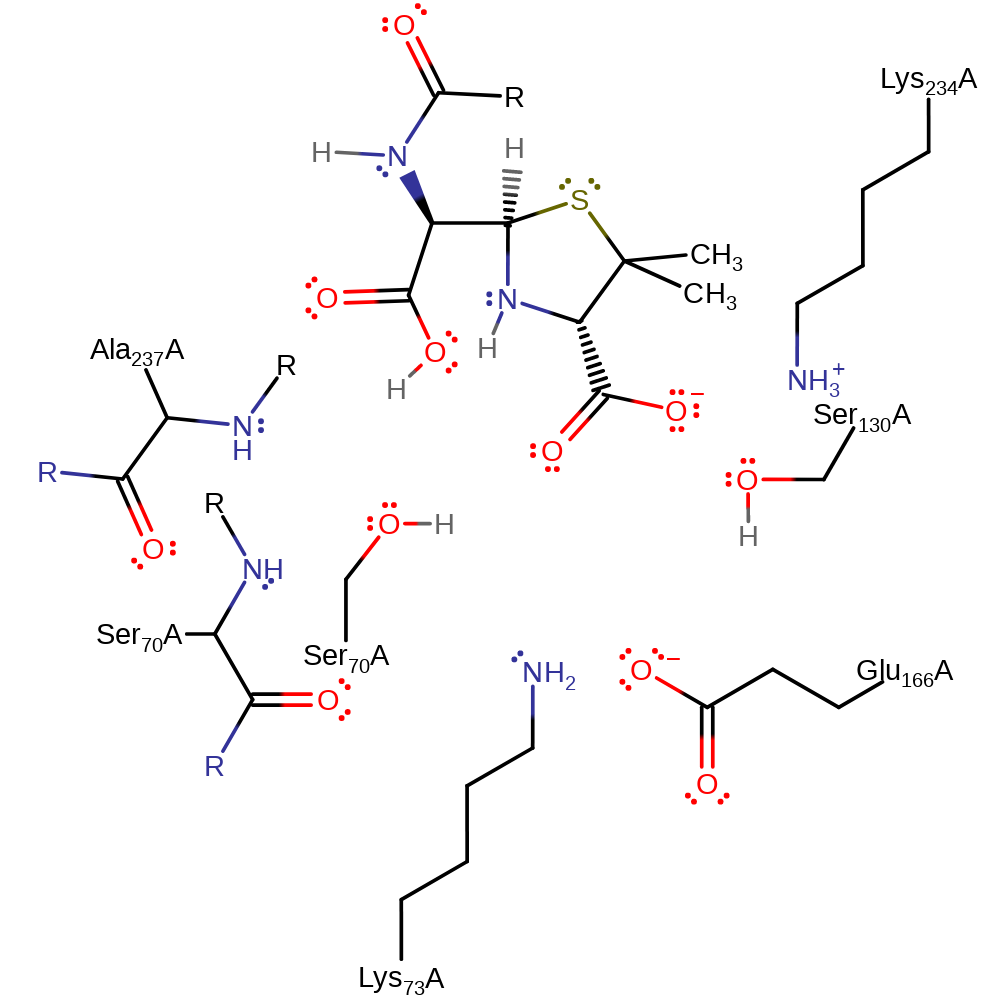 Download:
Download: