Glycine amidinotransferase
Glycine amidinotransferase, also known as L-arginine:glycine amidinotransferase (AT) catalyses the committed step in creatine biosynthesis. The enzyme does this by forming guanidinoacetic acid, the immediate precursor of creatine. Both creatine and its phosphorylated form play an essential role in the energy metabolism of muscle and nerve tissues, acting as a dynamic reservoir of high-energy phosphate which buffers the rapid fluctuations of the ATP/ADP ratio during muscle and nerve action.
Reference Protein and Structure
- Sequence
-
P50440
 (2.1.4.1)
(2.1.4.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1jdw
- CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS
(1.9 Å)



- Catalytic CATH Domains
-
3.75.10.10
 (see all for 1jdw)
(see all for 1jdw)
Enzyme Reaction (EC:2.1.4.1)
Enzyme Mechanism
Introduction
The reaction comprises of two steps, both involving nucleophilic attack. The first step starts by the addition of the carbon atom of the guanidino group of L-Arg to the thiol group of Cys407 of the enzyme, and donation of the thiol proton to the L-Arg substrate. The thiol group of C407 forms a covalently bound intermediate with the carbon atom of the guanidino group, and donates its proton to the imino group of the arginine substrate. Hydrogen bonds and salt linkage of the guanidino nitrogens to Asp170 and Asp305 enhance the electrophilicity of the guanidino-carbon atom, and a potential hydrogen bond of the epsilon-imino nitrogen to His303 supports proton transfer to this atom. Subsequently the bond between the epsilon-imino group and the amidino-carbon atom is broken to generate L-Orn. The second step starts with a proton transfer from the positively charged Gly substrate to His303 and a nucleophilic attack of the lone electron pair of the Gly nitrogen atom at the carbon atom of the bound amidino group. This is followed by the formation of a tetrahedral adduct and its collapse by cleavage of the amidino-carbon-sulfur bond to generate guanidinoacetic acid.
The amidino-carbon atom of the arginine substrate is placed between the thiol group of the cysteine residue and the imino group of the histidine residue. The arginine is fixed tightly to the active site by formation of hydrogen bonds between the amidino nitrogen atoms and the carboxyl groups of aspartate residues 170 and 305. These orient the substrate's guanidino group such that the sulphur atom of CYS 407, the guanidino-carbon atom and the nitrogen atom of HIS 303 are in a line orthogonal to the plane of the guanidino group.
Catalytic Residues Roles
| UniProt | PDB* (1jdw) | ||
| Asp254 | Asp254A | Forms part of the charge relay Cys-His-Asp catalytic triad. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Cys407 | Cys407A | Acts as the nucleophile in the initial step of the reaction. It is activated through a Cys-His-Asp catalytic triad. | hydrogen bond acceptor, hydrogen bond donor, nucleophile, proton acceptor, proton donor, nucleofuge |
| His303 | His303A | Forms part of the charge relay Cys-His-Asp catalytic triad. Acts as a general acid/base, abstracting a proton from amine of the substrate glycine and is returned to its initial protonation state by a water molecule in an inferred return step. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asp170, Asp305 | Asp170A, Asp305A | Helps stabilise the intermediates formed during the course of the reaction. | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, overall product formed, intermediate formation, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminated, native state of enzyme regenerated, inferred reaction stepReferences
- Shirai H et al. (2001), Trends Biochem Sci, 26, 465-468. A novel superfamily of enzymes that catalyze the modification of guanidino groups. DOI:10.1016/s0968-0004(01)01906-5. PMID:11504612.
- Tsikas D et al. (2015), Amino Acids, 47, 1697-1702. Homoarginine, arginine, and relatives: analysis, metabolism, transport, physiology, and pathology. DOI:10.1007/s00726-015-2055-5. PMID:26210755.
- Fritsche E et al. (1999), J Biol Chem, 274, 3026-3032. The Ligand-induced Structural Changes of HumanL-Arginine:Glycine Amidinotransferase: A MUTATIONAL AND CRYSTALLOGRAPHIC STUDY. DOI:10.1074/jbc.274.5.3026.
- Humm A et al. (1997), EMBO J, 16, 3373-3385. Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis. DOI:10.1093/emboj/16.12.3373. PMID:9218780.
- Fritsche E et al. (1997), Eur J Biochem, 247, 483-490. Substrate Binding and Catalysis by L-arginine: Glycine Amidinotransferase - A Mutagenesis and Crystallographic Study. DOI:10.1111/j.1432-1033.1997.00483.x.
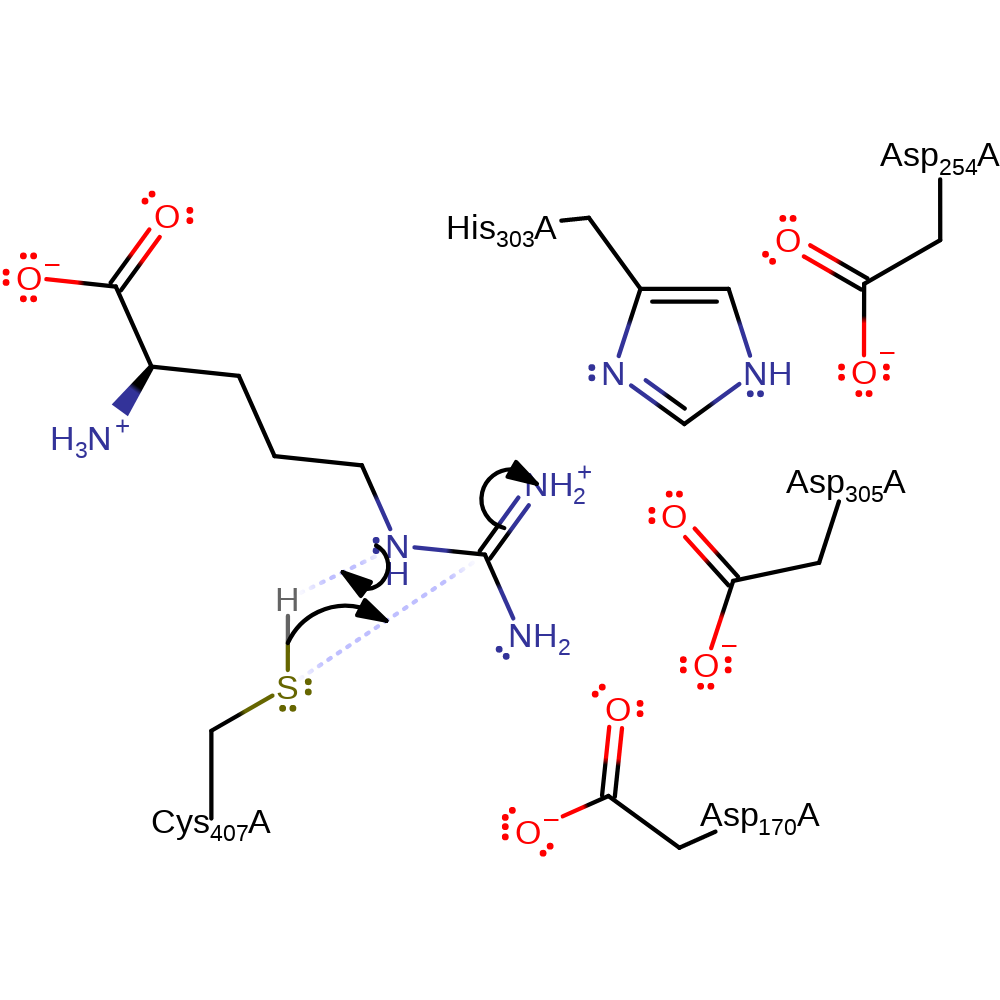
Step 1. The substrate arginine deprotonates Cys407, which initiates a nucleophilic attack on the guanido carbon of the arginine in a substitution reaction which eliminates ornithine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His303A | hydrogen bond donor, hydrogen bond acceptor |
| Asp305A | hydrogen bond acceptor, electrostatic stabiliser |
| Asp170A | hydrogen bond acceptor, electrostatic stabiliser |
| Cys407A | hydrogen bond donor |
| Asp254A | hydrogen bond acceptor |
| Cys407A | nucleophile, proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, overall product formed, intermediate formation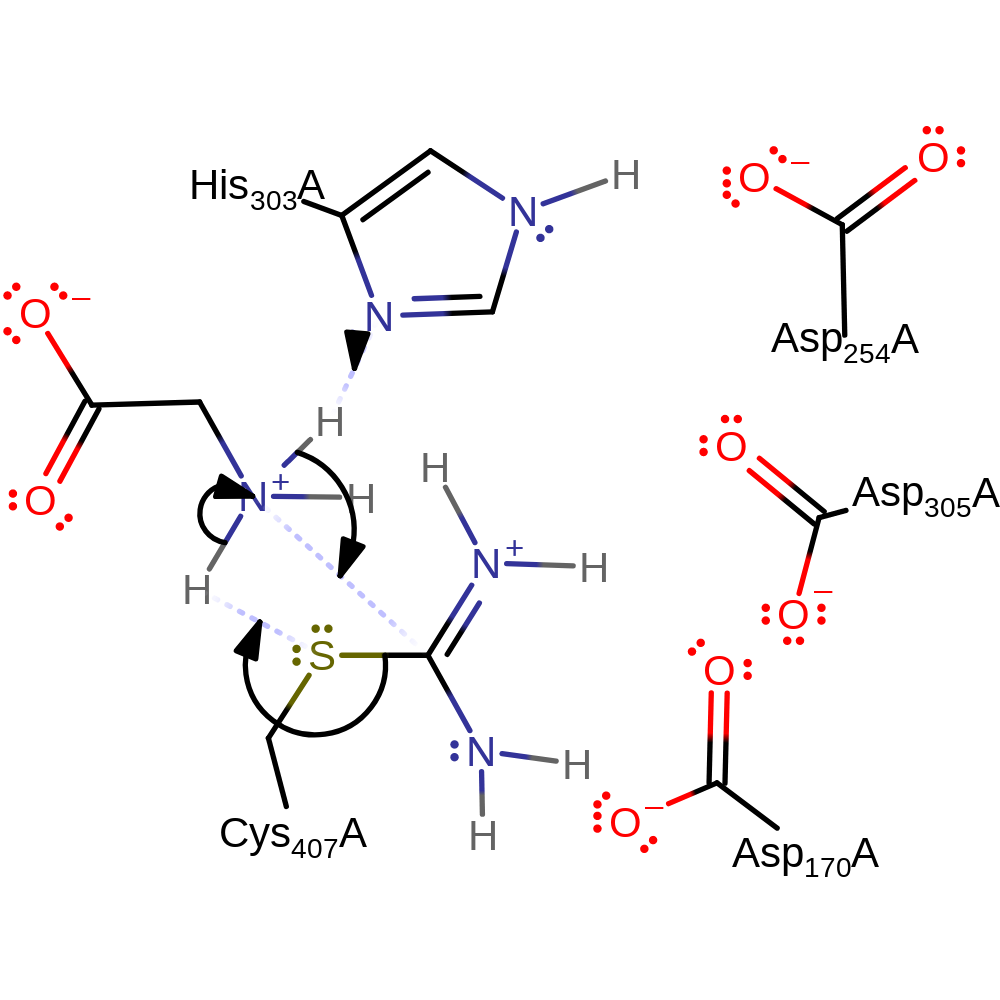
Step 2. His303 deprotonates the amine of the substrate glycine, which initiates a nucleophilic attack on the covalently bound carbon in a substitution reaction, eliminating Cys407, which deprotonates the substrate glycine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His303A | hydrogen bond acceptor, hydrogen bond donor |
| Asp305A | hydrogen bond acceptor, electrostatic stabiliser |
| Asp170A | hydrogen bond acceptor, electrostatic stabiliser |
| Asp254A | hydrogen bond acceptor, activator |
| Cys407A | hydrogen bond acceptor, proton acceptor, nucleofuge |
| His303A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, enzyme-substrate complex cleavage, intermediate collapse, intermediate terminatedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His303A | hydrogen bond donor |
| Asp254A | hydrogen bond acceptor, electrostatic stabiliser |
| His303A | proton donor |




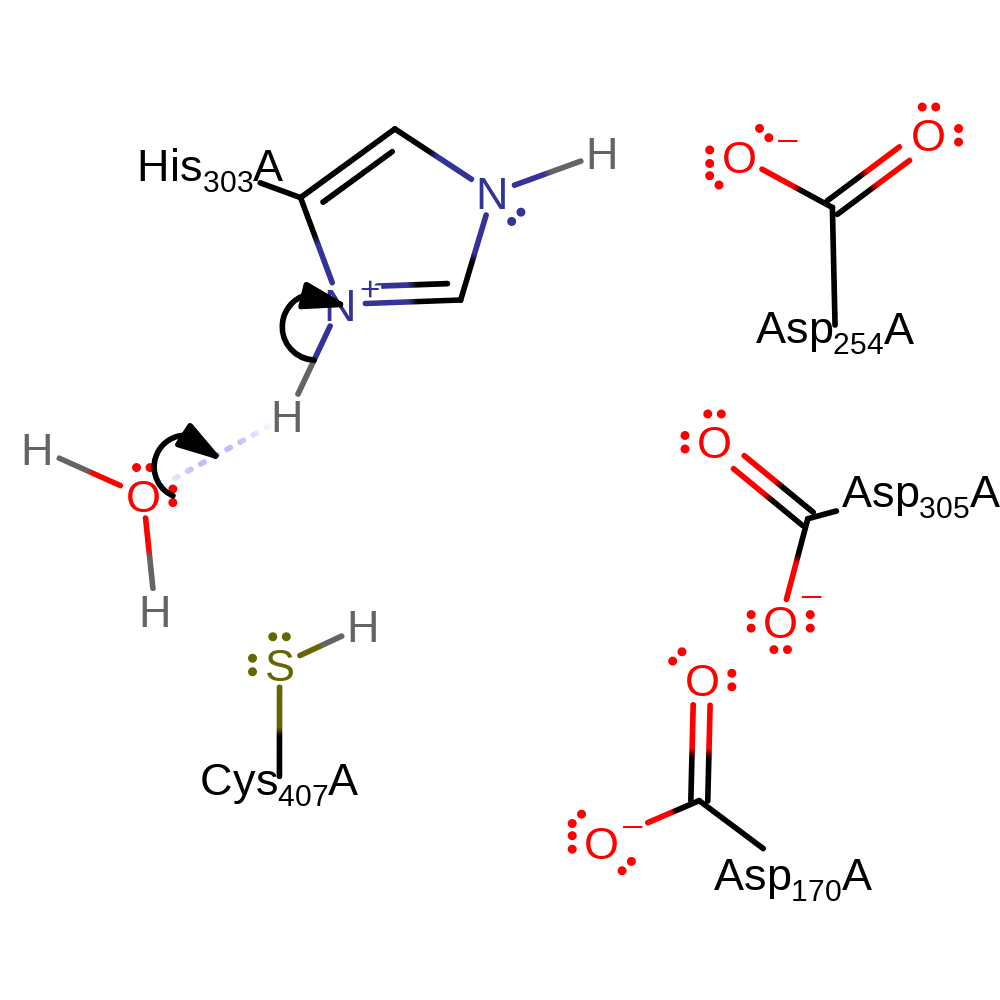
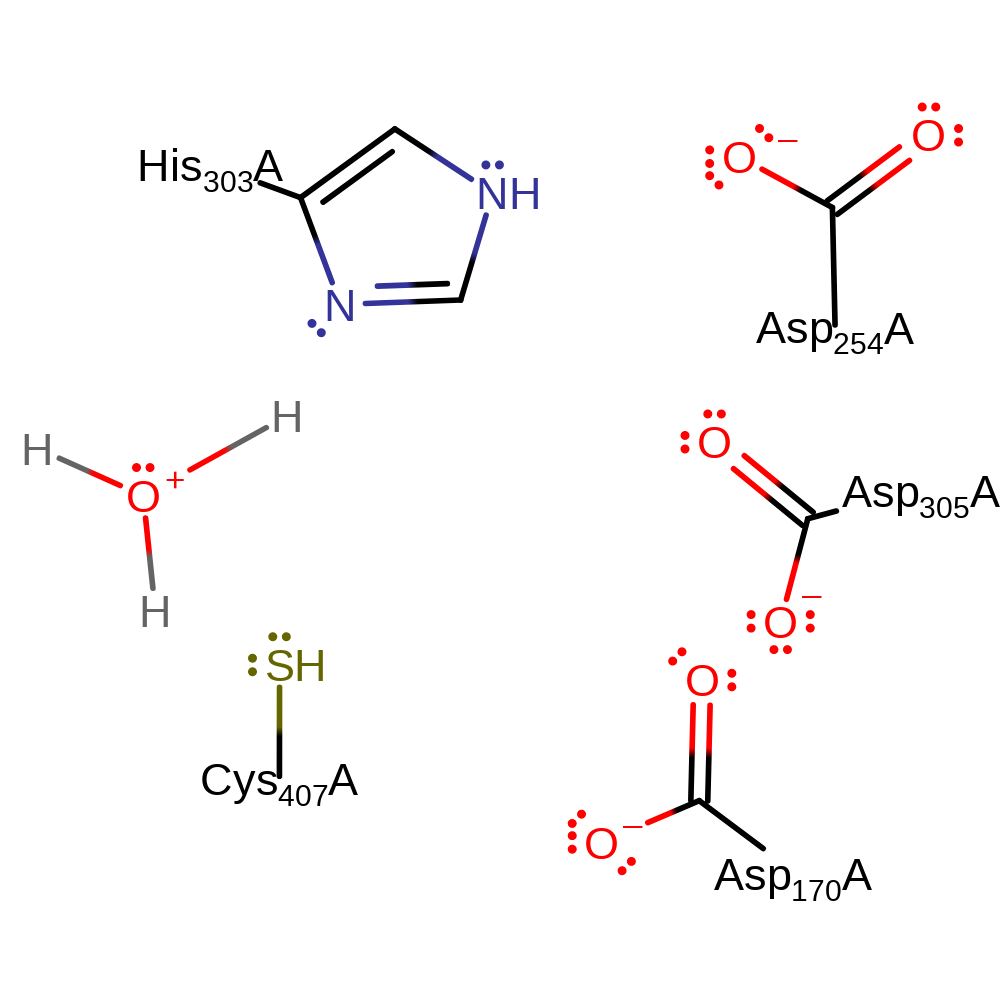 Download:
Download: