Protein-methionine-S-oxide reductase (MsrA)
Peptide methionine sulfoxide reductase (MsrA) reverses the oxidative damage to both free methionine and methionine within a protein by catalysing the metal/cofactor independent reduction of the sulphoxide moiety. The reducing equivalents are provided by either DTT or a thioredoxin-regenerating system. The structure of MrsA is of a mixed alpha/beta type, with a two-layer alpha-beta sandwich motif at its core.
Methionine oxidation is a serious problem for cells. It can lead to inviability of the cells if it goes on unchecked, and even in systems where MsrA s present methionine oxidation is still a major cause of random cell damage and has been implicated in a variety of neuro-generative deceases, emphysema, cataractogenesis, rheumatoid arthritis and other general ageing processes.
Reference Protein and Structure
- Sequence
-
P54149
 (1.8.4.11)
(1.8.4.11)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bos taurus (Cattle)

- PDB
-
1fva
- CRYSTAL STRUCTURE OF BOVINE METHIONINE SULFOXIDE REDUCTASE
(1.7 Å)



- Catalytic CATH Domains
-
3.30.1060.10
 (see all for 1fva)
(see all for 1fva)
Enzyme Reaction (EC:1.8.4.11)
Enzyme Mechanism
Introduction
In this proposal, the oxygen atom on the sulfenic acid is derived from a water molecule rather than methionine sulfoxide. Computational studies suggest that this proposal is the more likely.
The reaction is initiated by Glu115 donating a proton to the sulfoxide, and the protonated hydroxyl sulfonium intermediate is stabilised by hydrogen bonding with Tyr103 and Tyr155.Nucleophilic attack of the hydroxyl sulfonium intermediate by the thiolate of Cys72 forms the sulfurane intermediate. Tyr103 then donates a proton to the hydroxyl sulfonium intermediate, at which point a water molecule is eliminated. Another water molecule, seen in several crystal structures of MsrA is sited 2.8 Å from Cys72 on the opposite side from the bound substrate. This location places it in perfect position to react with the sulfonium intermediate to form the sulfenic acid on Cys72 and release Met as a product. After the release of Met, the Cys72 sulfenic acid form can be reduced back to the thiolate by reducing agents, including thioredoxin, the C-terminal resolving cysteines or DTT.
Catalytic Residues Roles
| UniProt | PDB* (1fva) | ||
| Cys72 | Cys72(60)A | The residue is deprotonated in the positively charged protein environment, allowing the terminal sulfoxide to act as a nucleophile towards the substrate sulfur-oxygen d-pi double bond. This forms a tetrahedral intermediate, which once protonated at the oxygen to form an oxonium leaving group, is attacked in an Sn2 manner at the Cys72 sulfur atom by an active site solvent molecule to form a sulfoxide. The sulfoxide is destroyed by the nucleophilic attack of Cys218 to form of a disulfide bridge between Cys72 and Cys218. The enzyme is restored for the next reaction through disulfide formation with Cys227 and the involvement of DTT or a thioredoxin-regenerating system. | covalently attached, nucleofuge, nucleophile, electrofuge, electrophile |
| Asp150, Tyr103, Glu115 | Asp150(138)A, Tyr103(91)A, Glu115(103)A | Act as general acid/bases. | proton relay, proton acceptor, proton donor |
| Tyr155 | Tyr155(143)A | Acts to help stabilise the reactive intermediates and transition states formed. | electrostatic stabiliser |
| Cys218, Cys227 | Cys218(206)A, Cys227(215)A | Acts as nucleophiles in the regeneration of the active site. | nucleofuge, nucleophile, proton acceptor, proton donor, proton relay, electrofuge, electrophile |
Chemical Components
bimolecular nucleophilic addition, proton transfer, enzyme-substrate complex formation, elimination (not covered by the Ingold mechanisms), dehydration, bimolecular nucleophilic substitution, proton relay, enzyme-substrate complex cleavage, bimolecular homolytic substitution, inferred reaction step, native state of enzyme regeneratedReferences
- Dokainish HM et al. (2013), Biochemistry, 52, 1814-1827. A Molecular Dynamics and Quantum Mechanics/Molecular Mechanics Study of the Catalytic Reductase Mechanism of Methionine Sulfoxide Reductase A: Formation and Reduction of a Sulfenic Acid. DOI:10.1021/bi301168p. PMID:23418817.
- Lim JC et al. (2011), Proc Natl Acad Sci U S A, 108, 10472-10477. Methionine sulfoxide reductase A is a stereospecific methionine oxidase. DOI:10.1073/pnas.1101275108. PMID:21670260.
- Balta B et al. (2006), J Phys Chem A, 110, 7628-7636. Theoretical Study of the Reduction Mechanism of Sulfoxides by Thiols. DOI:10.1021/jp0573036. PMID:16774207.
- Antoine M et al. (2003), J Biol Chem, 278, 45352-45357. Kinetic Characterization of the Chemical Steps Involved in the Catalytic Mechanism of Methionine Sulfoxide Reductase A from Neisseria meningitidis. DOI:10.1074/jbc.m307471200. PMID:12954610.
- Boschi-Muller S et al. (2000), J Biol Chem, 275, 35908-35913. A Sulfenic Acid Enzyme Intermediate Is Involved in the Catalytic Mechanism of Peptide Methionine Sulfoxide Reductase fromEscherichia coli. DOI:10.1074/jbc.m006137200. PMID:10964927.
- Tête-Favier F et al. (2000), Structure, 8, 1167-1178. Crystal Structure of the Escherichia coli Peptide Methionine Sulphoxide Reductase at 1.9 Å Resolution. DOI:10.1016/s0969-2126(00)00526-8. PMID:11080639.
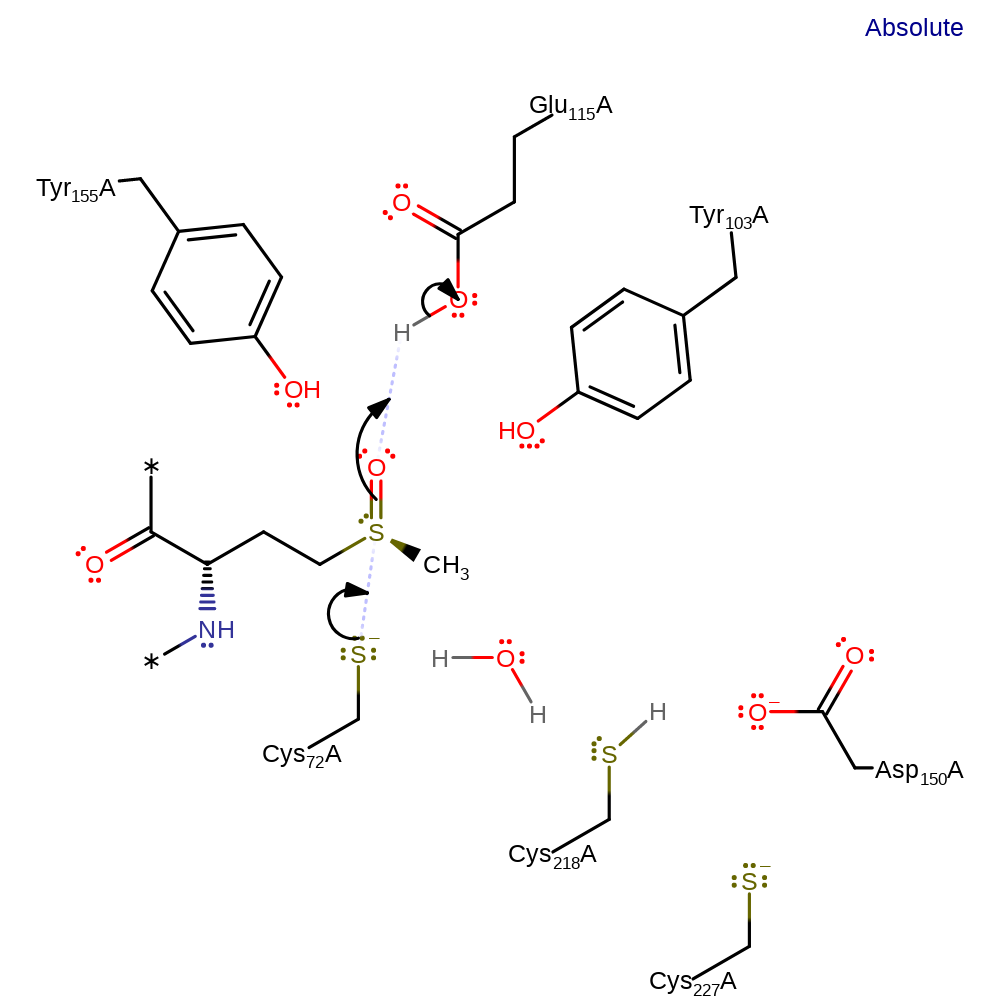
Step 1. Cys72 initiates a nucleophilic attack on the sulfoxide group, which abstracts a proton from Glu115.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr103(91)A | electrostatic stabiliser |
| Tyr155(143)A | electrostatic stabiliser |
| Cys72(60)A | nucleophile |
| Glu115(103)A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, enzyme-substrate complex formation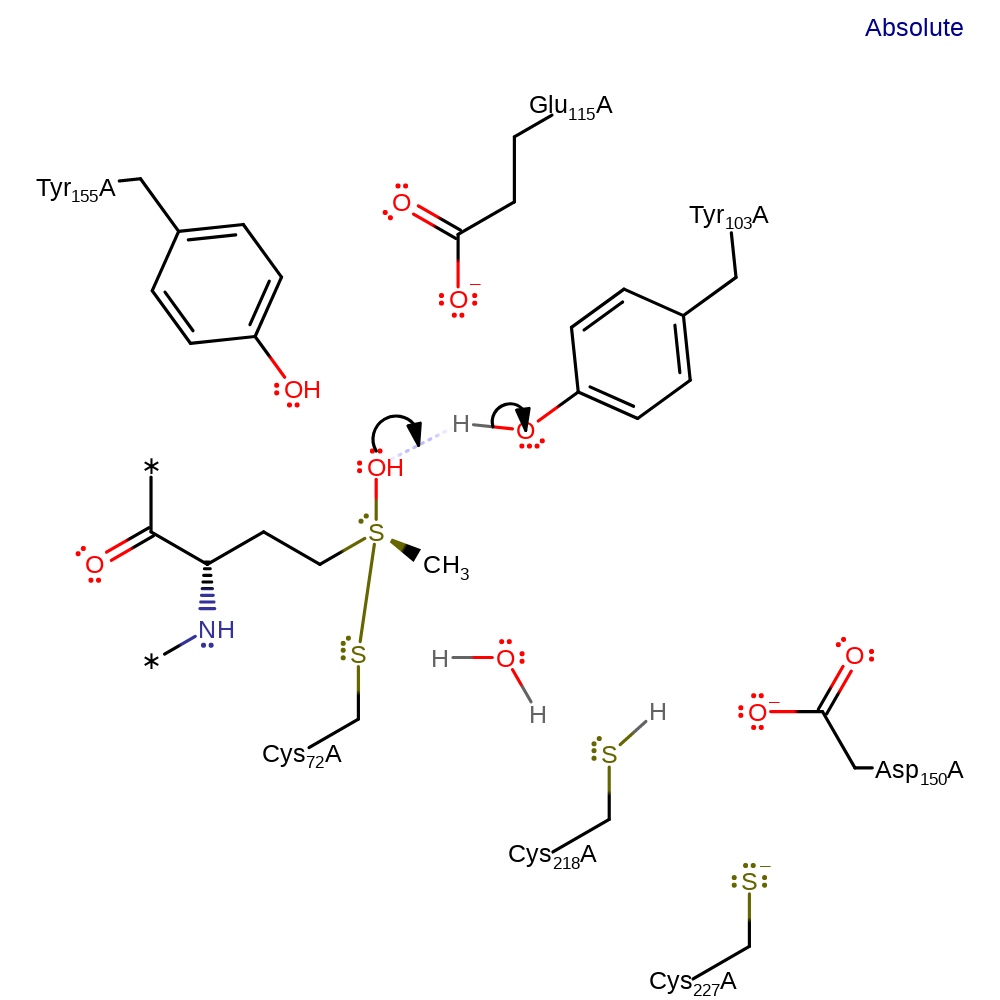
Step 2. The new hydroxyl group abstracts a proton from Tyr103, making it a better leaving group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr155(143)A | electrostatic stabiliser |
| Tyr103(91)A | proton donor |
Chemical Components
proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys72(60)A | covalently attached |
| Tyr103(91)A | electrostatic stabiliser |
| Tyr155(143)A | electrostatic stabiliser |
| Glu115(103)A | electrostatic stabiliser |
Chemical Components
elimination (not covered by the Ingold mechanisms), dehydration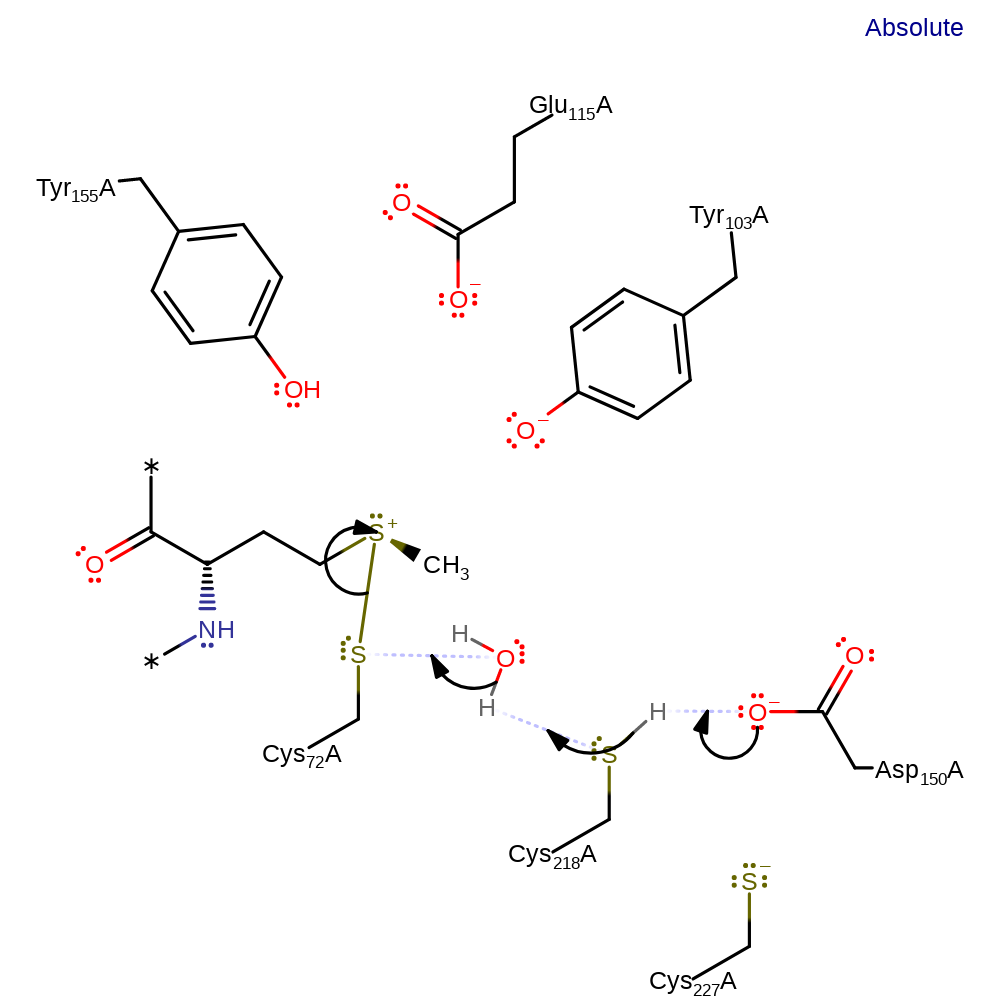
Step 4. An active site water, activated through a proton relay involving Asp150 and Cys218 initiates a nucleophilic attack on the sulfur of Cys72, eliminating the methionine product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys218(206)A | proton acceptor |
| Cys72(60)A | electrofuge |
| Cys218(206)A | proton relay, proton donor |
| Cys72(60)A | electrophile |
| Asp150(138)A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton relay, enzyme-substrate complex cleavage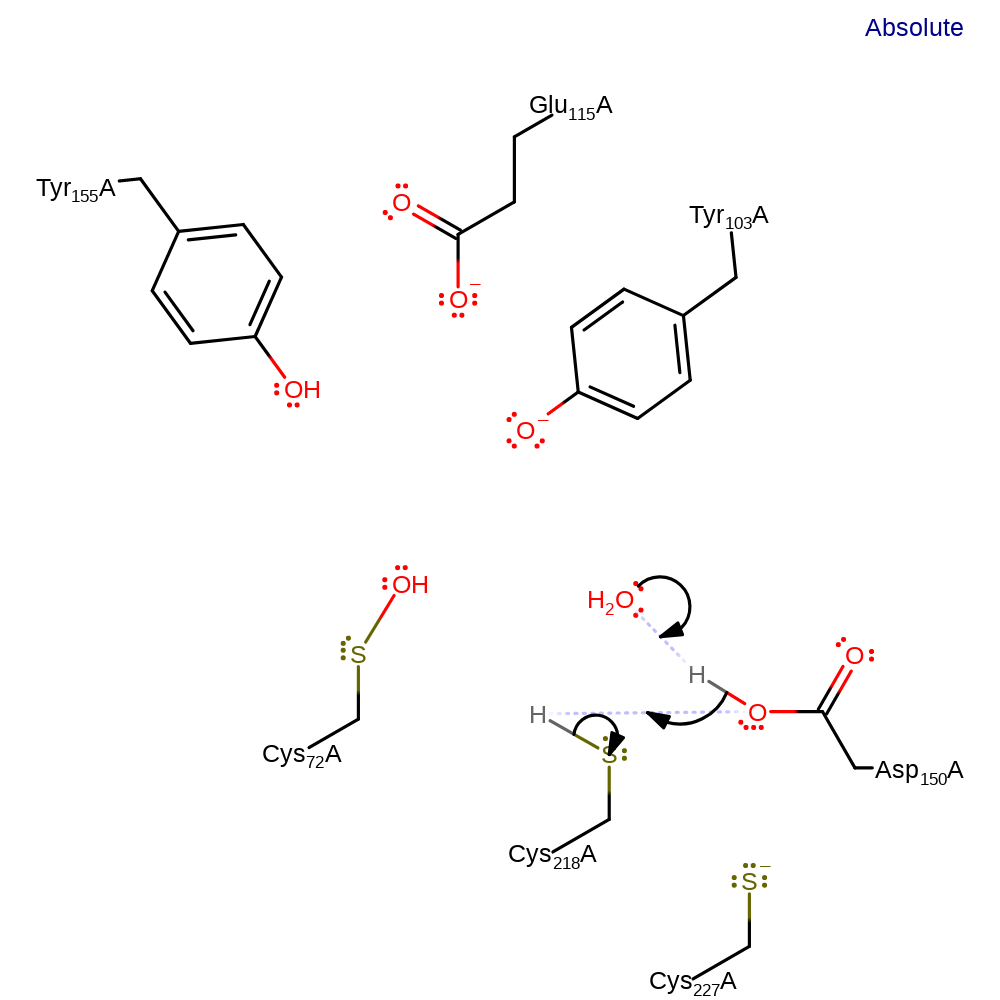
Step 5. Another water molecule abstracts a proton from Asp150, which in turn abstracts the proton from Cys218.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp150(138)A | proton acceptor, proton relay, proton donor |
| Cys218(206)A | proton donor |
Chemical Components
proton relay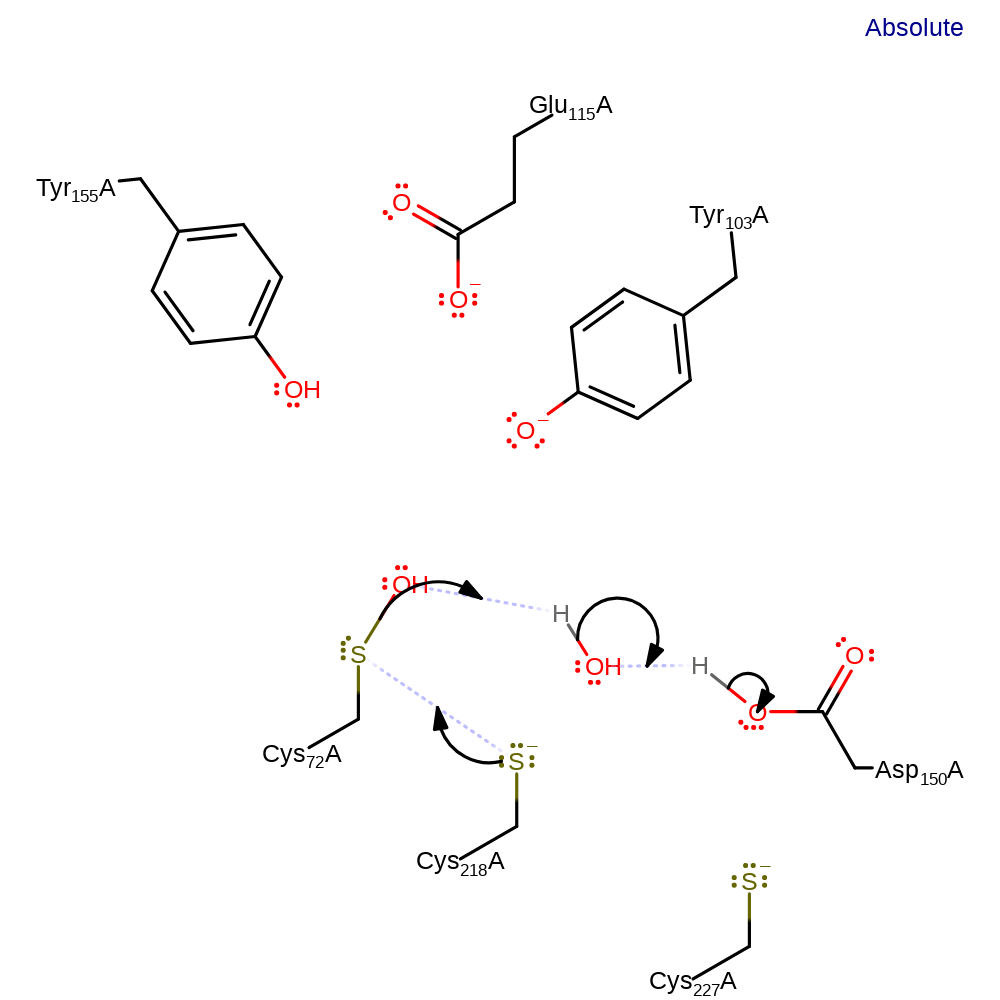
Step 6. The activated Cys218 attacks Cys72, eliminating a water molecule, which in turn abstracts a proton from Asp150.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp150(138)A | proton donor |
| Cys218(206)A | nucleophile |
| Cys72(60)A | electrophile, electrofuge |
Chemical Components
proton relay, ingold: bimolecular nucleophilic substitution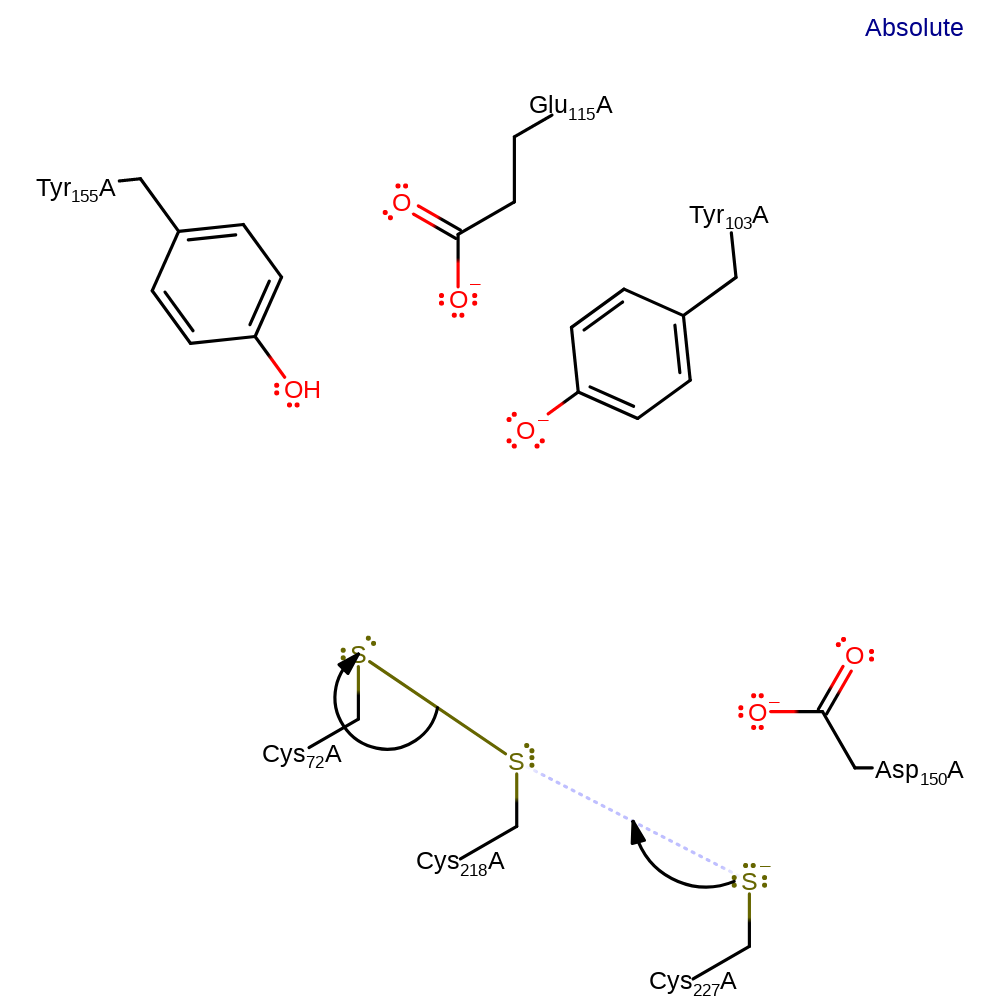
Step 7. Cys227 initiates a nucleophilic attack on Cys218 in a substitution reaction, eliminating Cys72.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys72(60)A | nucleofuge |
| Cys227(215)A | nucleophile |
| Cys218(206)A | electrofuge, electrophile |
Chemical Components
ingold: bimolecular homolytic substitution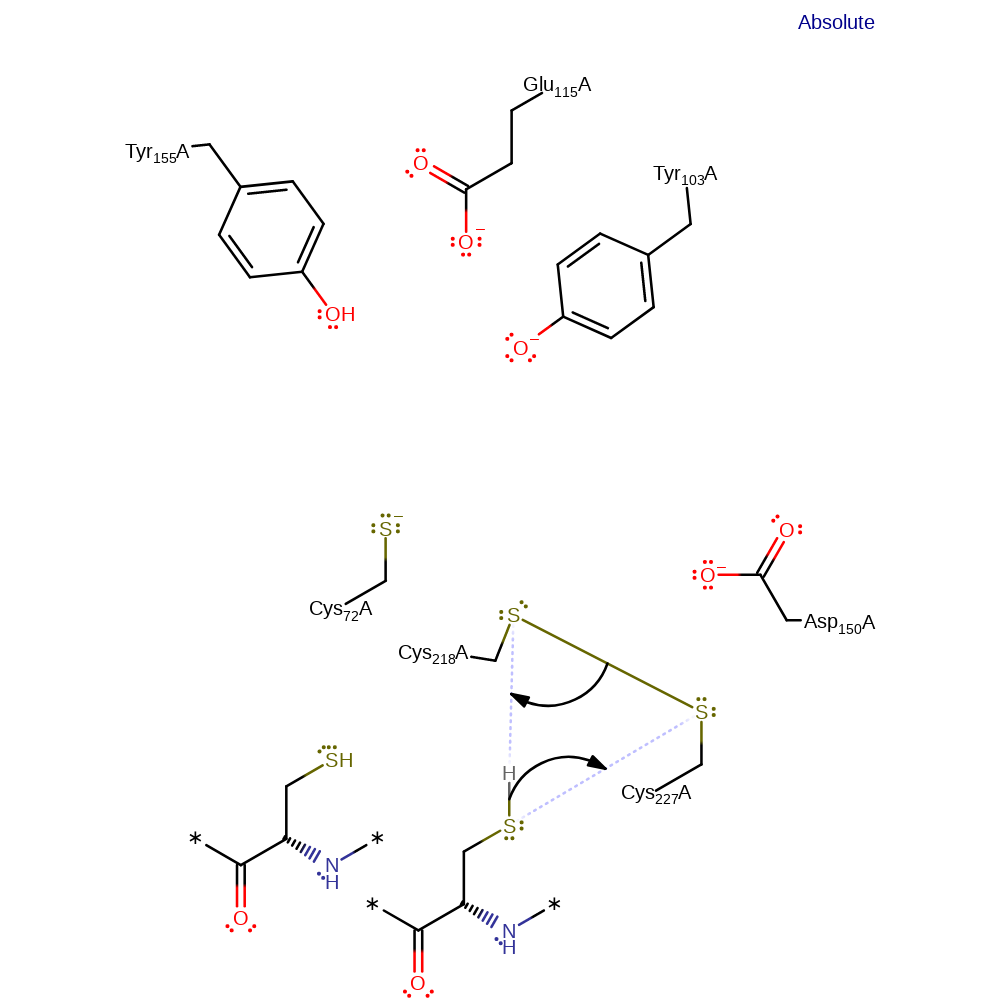
Step 8. Cys218 deprotonates one of the thiols of thioredoxin, which initiates a nucleophilic attack on Cys227 in a substitution reaction, eliminating Cys218.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys218(206)A | nucleofuge |
| Cys227(215)A | electrofuge |
| Cys218(206)A | proton acceptor |
| Cys227(215)A | electrophile |
Chemical Components
proton transfer, enzyme-substrate complex formation, ingold: bimolecular homolytic substitution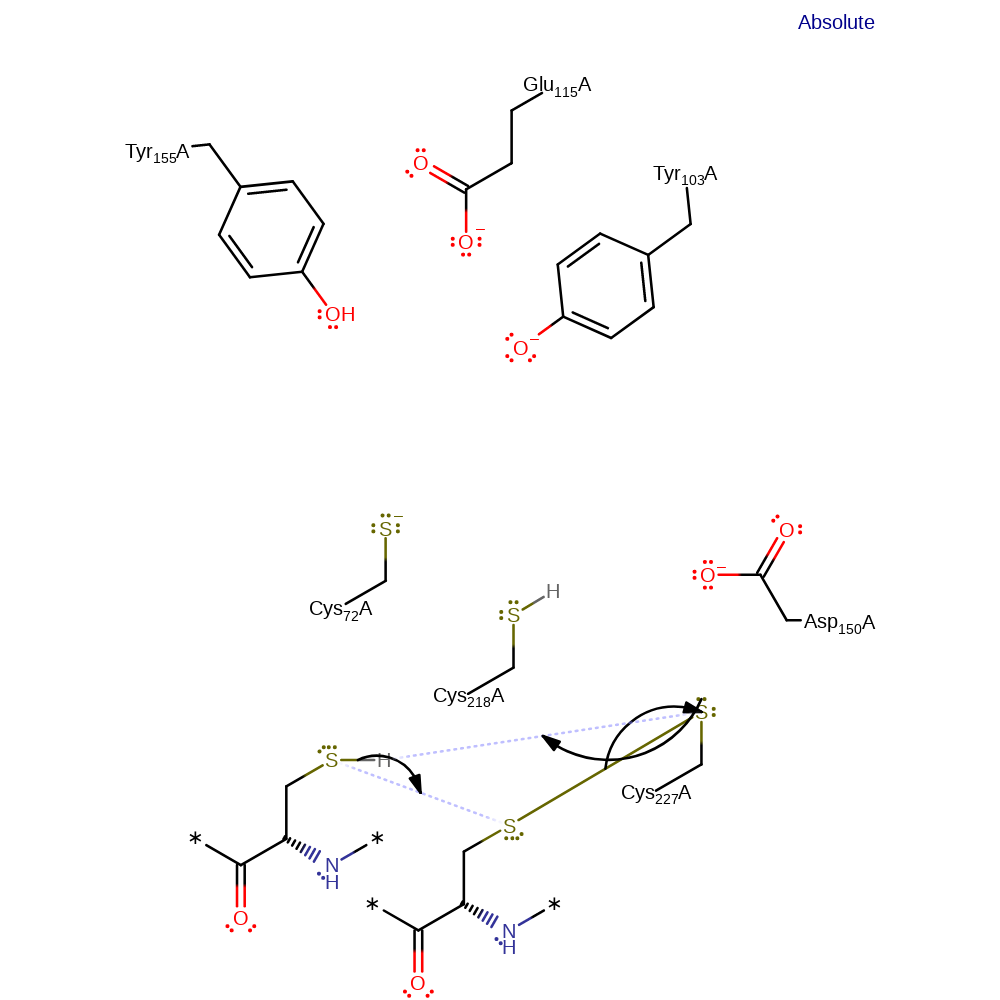
Step 9. Cys227 deprotonates the second thiol of thioredoxin, which initiates a nucleophilic attack on the sulfur of the thioredoxin that is covalently attached to Cys227 in a substitution reaction, eliminating Cys227.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys227(215)A | proton acceptor, nucleofuge |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, enzyme-substrate complex cleavage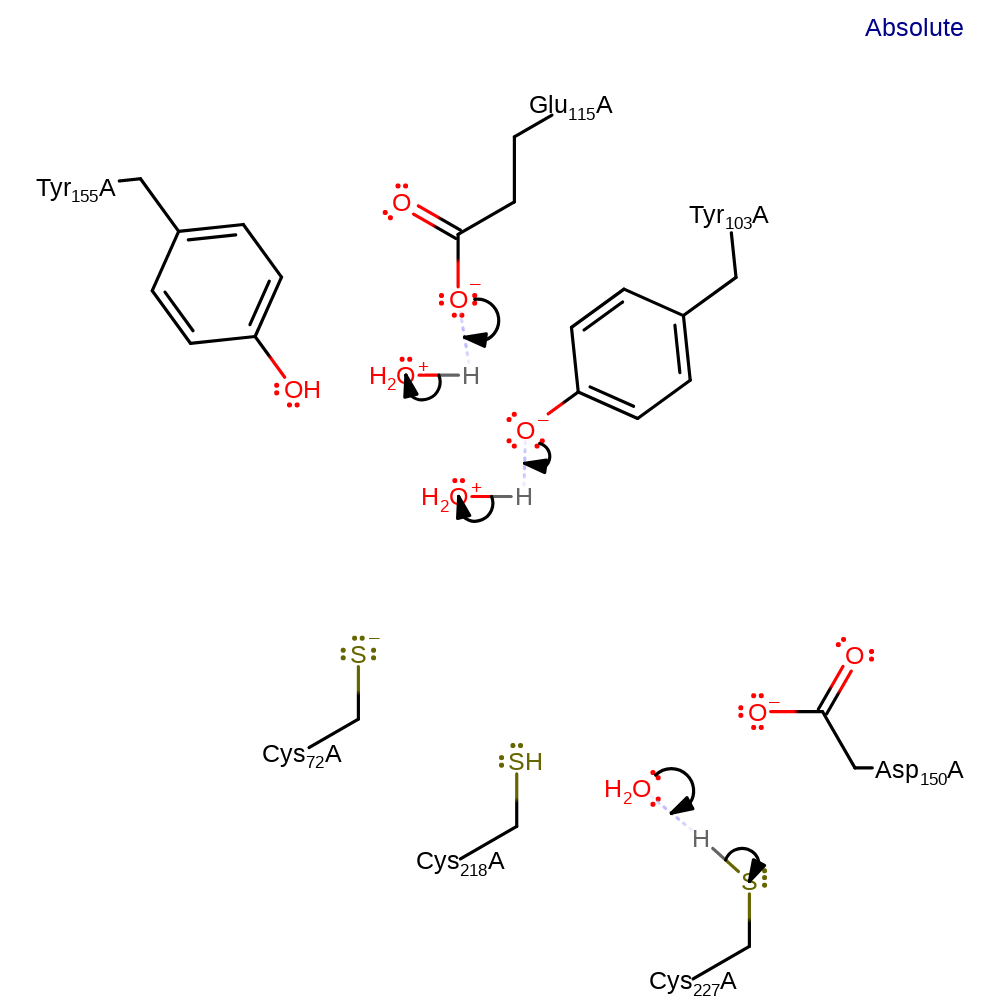
Step 10. Inferred return step to regenerate the starting protonation states of the active site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu115(103)A | proton acceptor |
| Cys227(215)A | proton donor |
| Tyr103(91)A | proton acceptor |
Chemical Components
proton transfer, inferred reaction step, native state of enzyme regeneratedIntroduction
In this proposal, direct formation of the Cys218-Cys72 bond results in the elimination of methionine and water.
Cys72 is activated and stabilised as the thiolate anion due to its located at the positive end of the alpha helix dipole. Nuceleophilic attack on the sulfur atom of Met(O) by Cys72 may be encouraged by simultaneous proton transfer from Glu115. The resulting tetravalent sulfur intermediate is stabilised by interactions with Tyr103, Glu115 ad Tyr115. A proton is transfer from Cys218 to the intermediate. This allows Cys218 to act as the second nucleophile at Cys72, resulting in the loss of water and the collapse of the intermediate to the sulphide form of methionine. The final stages of the reaction proceed via a thiol-disulfide exchange in which Cys227 nucleophilically attacks Cys218, breaking the disulfide bond between Cys227 and Cys72, and forming a new one between Cys218 and Cys227. Finally, either a DTT or a thioredoxin-regenerating system then restore the active site to its fully reduced state.
Catalytic Residues Roles
| UniProt | PDB* (1fva) | ||
| Cys218 | Cys218(206)A | The residue acts as a general acid towards the tetrahedral intermediate, protonating the hydroxide, and so forming a water leaving group. The resulting anionic residue side chain sulphoxide is then positioned to act as a nucleophile towards the intermediate, initiating the release of water and methionine and the formation of a disulphide bridge between Cys217 and Cys 72. | hydrogen bond donor, nucleophile, nucleofuge, proton acceptor, proton donor, electrofuge, electrophile |
| Asp150 | Asp150(138)A | This residue is thought to act as a general acid/base in the other mechanism proposal, and so the same function has been inferred in this proposal. | proton acceptor, proton donor |
| Cys72 | Cys72(60)A | The residue is deprotonated in the positively charged protein environment, allowing the terminal sulfoxide to act as a nucleophile towards the substrate sulfur-oxygen d-pi double bond. This forms a tetrahedral intermediate which, once protonated by Cys218 at the oxygen to form an oxonium leaving group, is attacked in an Sn2 manner at the Cys72 sulfur atom by the now sulfoxide Cys218. This results in the displacement of methionine and the formation of a disulfide bridge between Cys72 and Cys218. The enzyme is restored for the next reaction through disulfide formation with Cys227 and the involvement of DTT or a thioredoxin-regenerating system. | covalently attached, nucleofuge, nucleophile, electrofuge, electrophile |
| Glu115 | Glu115(103)A | Acts as a general acid towards the substrate carbonyl during the nucleophilic attack of Cys72, increasing the substrate's electrophilicity and stabilising the alkoxide-character transition state. The residue is reprotonated by Cys227 on regeneration of the active site. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Tyr103, Tyr155 | Tyr103(91)A, Tyr155(143)A | The residue stabilises the anionic-character transition state through hydrogen bonding between the phenolic hydroxyl group and the intermediate's hydroxyl group. | proton acceptor, hydrogen bond donor, electrostatic stabiliser, proton donor |
| Cys227 | Cys227(215)A | Cys227 is deprotonated by Glu115 to restore the active site for catalysis. The resulting sulphoxide attacks the neighbouring Cys218, breaking its disulphide link to Cys72, and forming a new link to Cys218. Cys227 then acts as an electron acceptor in the reduction of this disulphide link by DDT or thioredoxin, leaving the active site ready to catalyse a further reaction. | hydrogen bond donor, nucleofuge, proton acceptor, proton donor, nucleophile, electrofuge, electrophile |
Chemical Components
bimolecular nucleophilic addition, proton transfer, enzyme-substrate complex formation, bimolecular nucleophilic substitution, enzyme-substrate complex cleavage, dehydration, native state of enzyme regenerated, inferred reaction stepReferences
- Lowther WT et al. (2000), Proc Natl Acad Sci U S A, 97, 6463-6468. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. DOI:10.1073/pnas.97.12.6463. PMID:10841552.
- Tête-Favier F et al. (2000), Structure, 8, 1167-1178. Crystal Structure of the Escherichia coli Peptide Methionine Sulphoxide Reductase at 1.9 Å Resolution. DOI:10.1016/s0969-2126(00)00526-8. PMID:11080639.
- Boschi-Muller S et al. (2000), J Biol Chem, 275, 35908-35913. A Sulfenic Acid Enzyme Intermediate Is Involved in the Catalytic Mechanism of Peptide Methionine Sulfoxide Reductase fromEscherichia coli. DOI:10.1074/jbc.m006137200. PMID:10964927.
- Lowther WT et al. (2000), Biochemistry, 39, 13307-13312. Structure and Mechanism of Peptide Methionine Sulfoxide Reductase, an “Anti-Oxidation” Enzyme†,‡. DOI:10.1021/bi0020269. PMID:11063566.
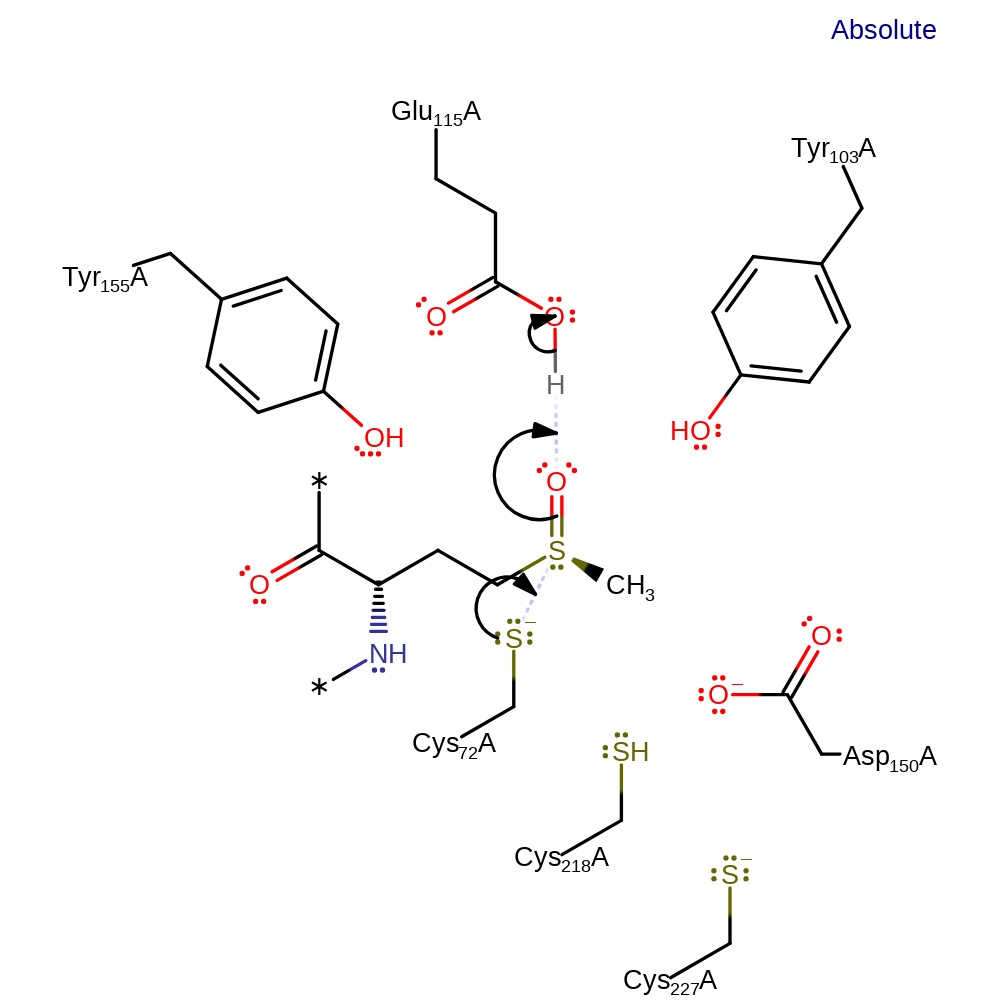
Step 1. Cys72 initiates a nucleophilic attack on the sulfoxide in an addition reaction with concomitant deprotonation of Glu115. Cys72 is activated and stabilised as a thiolate anion due to its location at the positive end of the alpha1 helix dipole.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr155(143)A | hydrogen bond donor, electrostatic stabiliser |
| Glu115(103)A | hydrogen bond donor |
| Tyr103(91)A | hydrogen bond donor, electrostatic stabiliser |
| Cys72(60)A | nucleophile |
| Glu115(103)A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, enzyme-substrate complex formation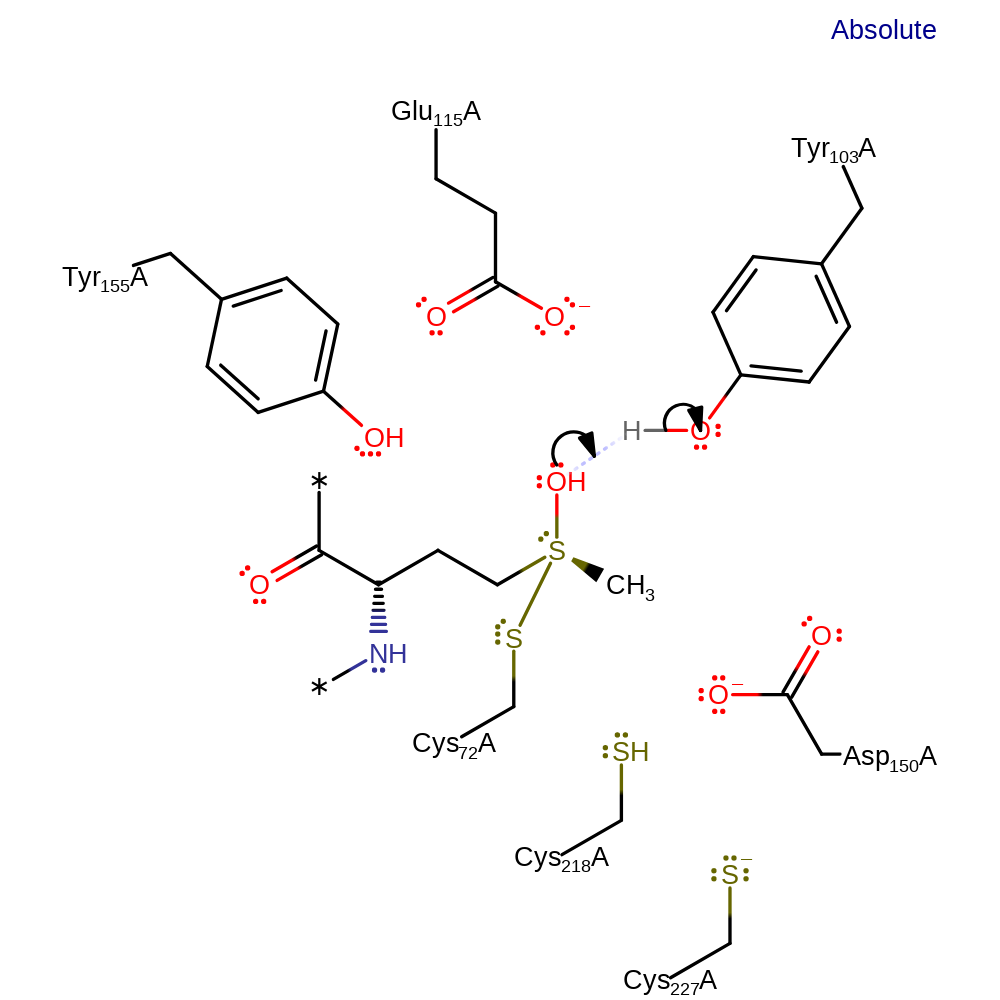
Step 2. The hydroxyl group of the intermediate abstracts a proton from Tyr103.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr155(143)A | hydrogen bond donor |
| Glu115(103)A | hydrogen bond acceptor |
| Tyr103(91)A | hydrogen bond donor |
| Cys72(60)A | covalently attached |
| Tyr103(91)A | proton donor |
Chemical Components
proton transfer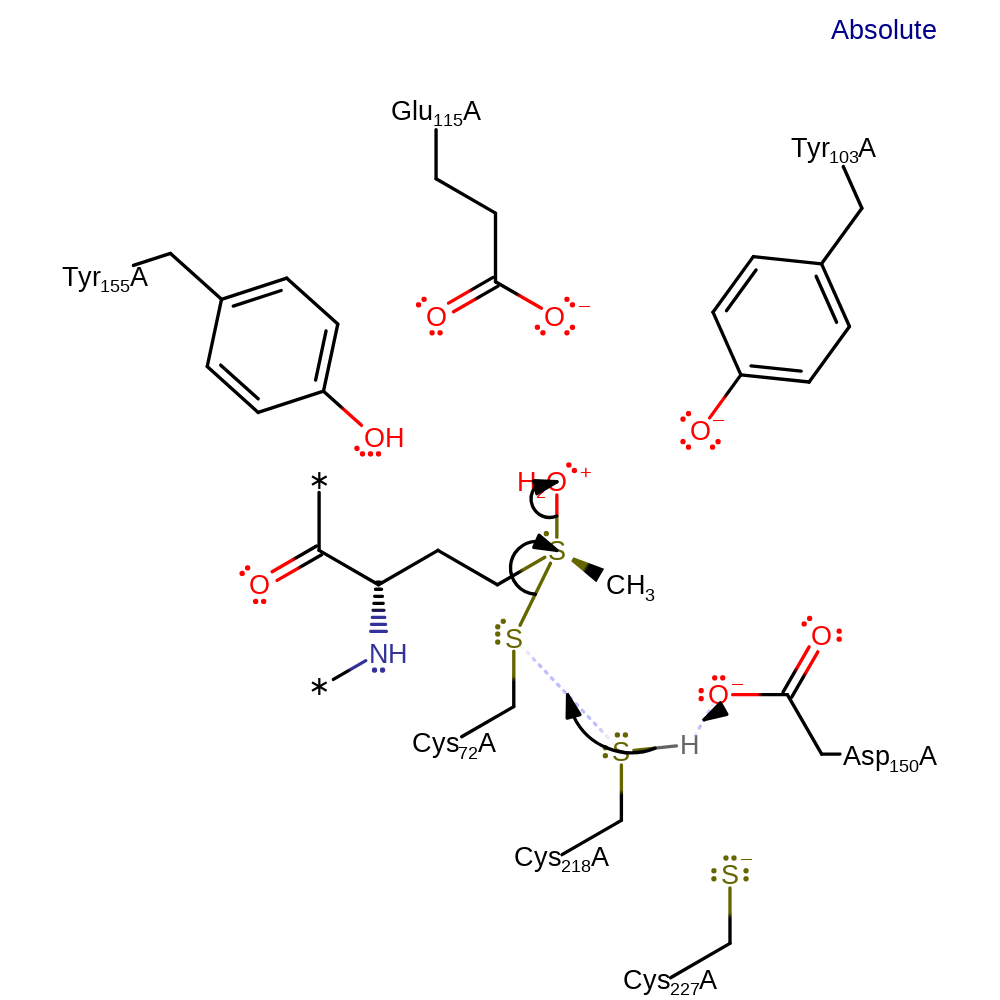
Step 3. Asp150 (inferred) abstracts the proton from Cys218, which initiates a nucleophilic attack on Cys72, eliminating water and methioinine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr155(143)A | hydrogen bond donor |
| Glu115(103)A | hydrogen bond acceptor |
| Tyr103(91)A | hydrogen bond donor |
| Cys218(206)A | hydrogen bond donor, nucleophile |
| Asp150(138)A | proton acceptor |
| Cys72(60)A | electrophile |
| Cys218(206)A | proton donor |
| Cys72(60)A | electrofuge |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, enzyme-substrate complex cleavage, dehydration
Step 4. Cys227 initiates a nucleophilic attack on Cys218 in a substitution reaction, eliminating Cys72.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys218(206)A | electrophile, electrofuge |
| Cys72(60)A | nucleofuge |
| Cys227(215)A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution
Step 5. Cys218 deprotonates one of the thiols of thioredoxin, which initiates a nucleophilic attack on Cys227 in a substitution reaction, eliminating Cys218.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys218(206)A | nucleofuge |
| Cys227(215)A | electrofuge |
| Cys218(206)A | proton acceptor |
| Cys227(215)A | electrophile |
Chemical Components
enzyme-substrate complex formation, proton transfer, ingold: bimolecular nucleophilic substitution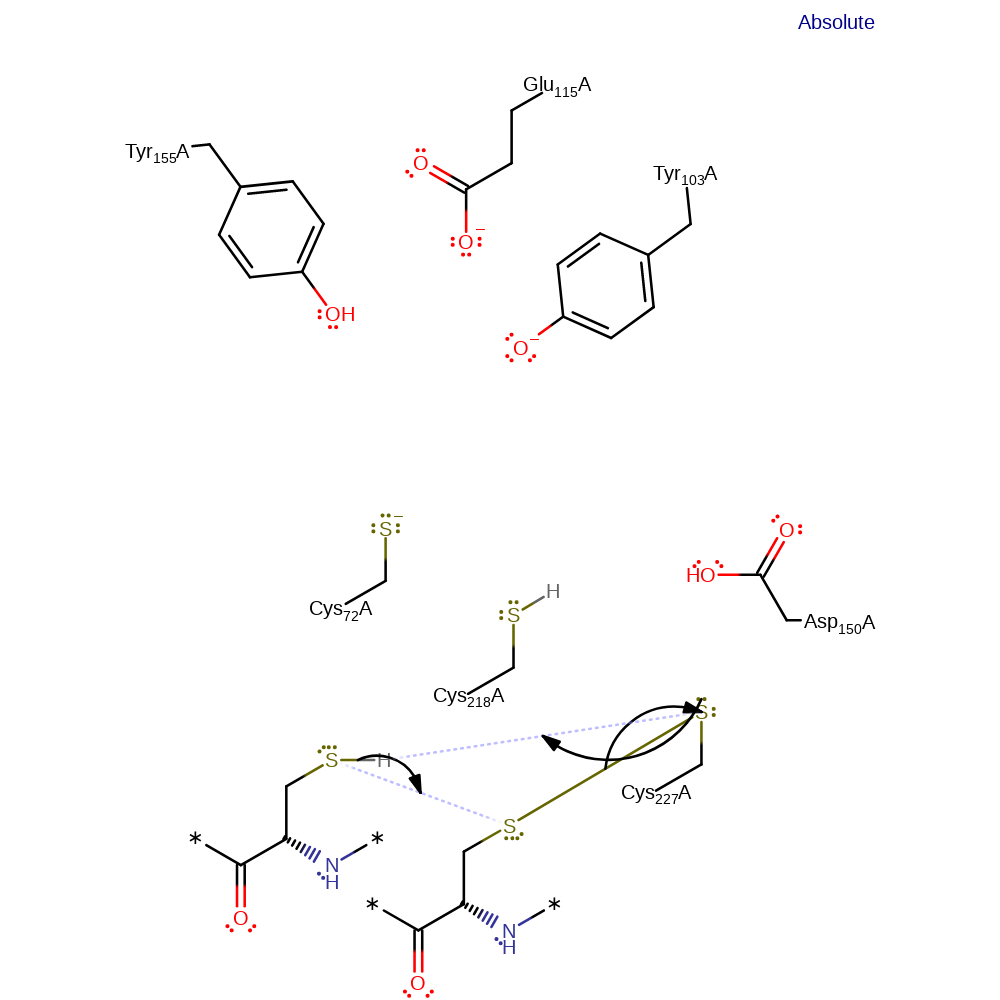
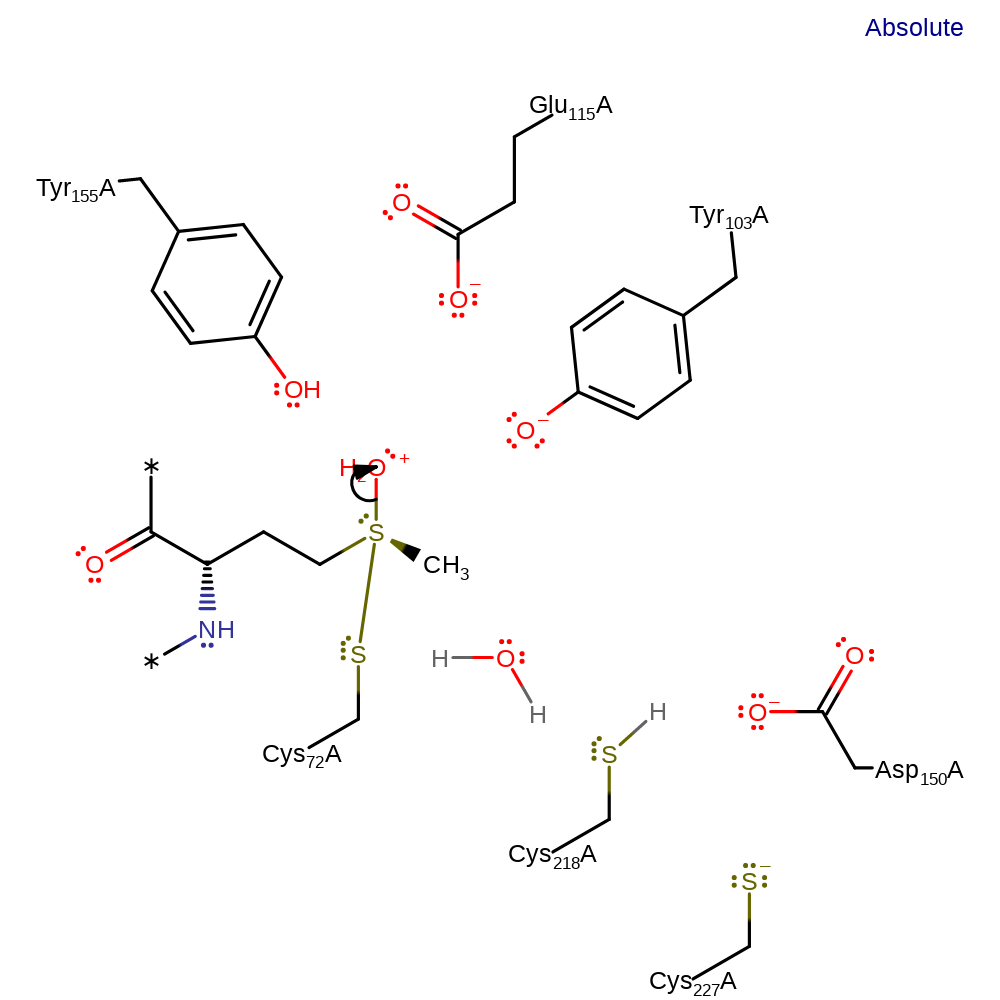
Step 6. Cys227 deprotonates the second thiol of thioredoxin, which initiates a nucleophilic attack on the sulfur of the thioredoxin that is covalently attached to Cys227 in a substitution reaction, eliminating Cys227.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys227(215)A | proton acceptor, nucleofuge |
Chemical Components
ingold: bimolecular nucleophilic substitution, enzyme-substrate complex cleavage, proton transfer
Step 7. In an inferred return step, several water molecules act as general acid/bases to return the active site to its starting protonation state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu115(103)A | hydrogen bond acceptor |
| Cys227(215)A | hydrogen bond donor |
| Glu115(103)A | proton acceptor |
| Asp150(138)A | proton donor |
| Cys227(215)A | proton donor |
| Tyr103(91)A | proton acceptor |





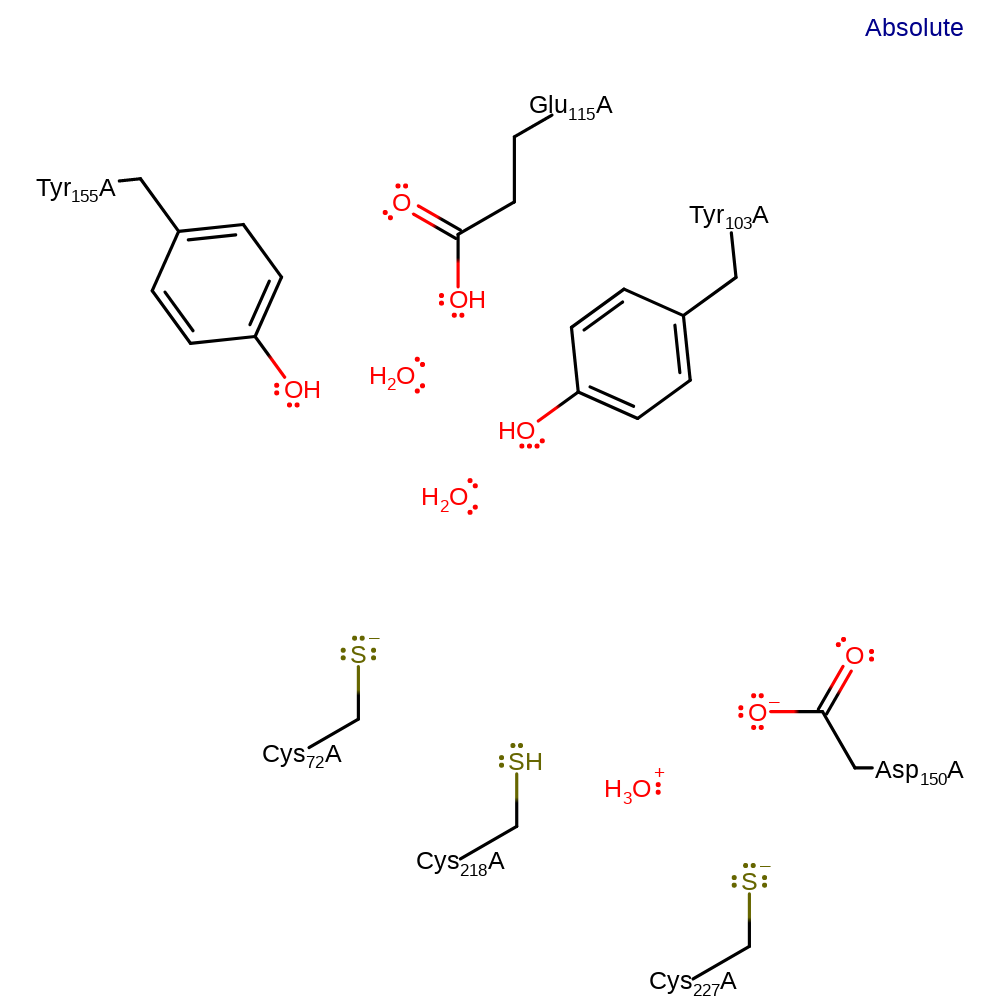 Download:
Download: 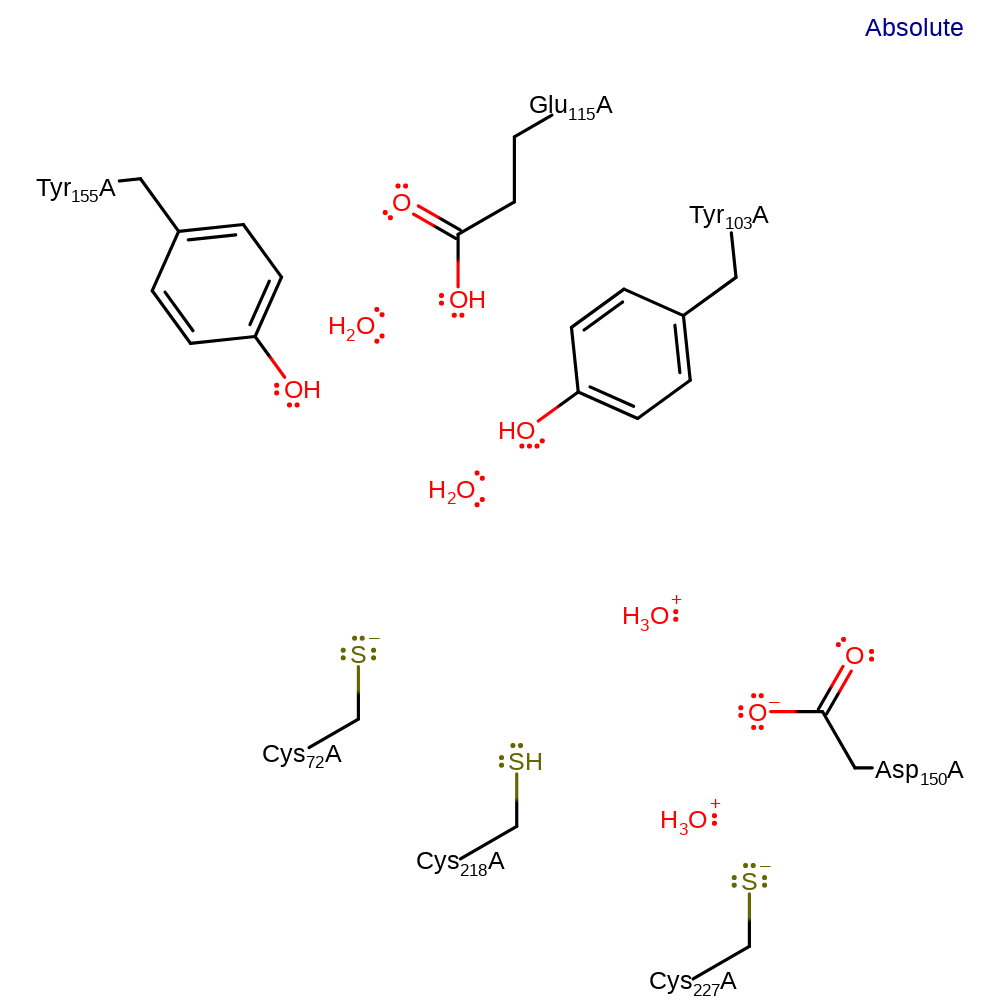 Download:
Download: