 |
PDBsum entry 5h3c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Immune system
|
PDB id
|
|

|

|
5h3c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Immune system
|
 |
|
Title:
|
 |
Crystal structure of arabidopsis snc1 tir domain
|
|
Structure:
|
 |
Protein suppressor of npr1-1, constitutive 1. Chain: a, b. Fragment: unp residues 17-190. Synonym: atsnc1,disease resistance rpp5-like protein. Engineered: yes
|
|
Source:
|
 |
Arabidopsis thaliana. Mouse-ear cress. Organism_taxid: 3702. Gene: snc1, bal, at4g16890, dl4475c, fcaall.51. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.225
|
R-free:
|
0.278
|
|
|
Authors:
|
 |
K.G.Hyun,J.M.Yoon,J.J.Song
|
|
Key ref:
|
 |
K.G.Hyun
et al.
(2016).
Crystal structure of Arabidopsis thaliana SNC1 TIR domain.
Biochem Biophys Res Commun,
481,
146-152.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
22-Oct-16
|
Release date:
|
07-Dec-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.3.2.2.6
- ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
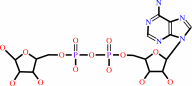 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochem Biophys Res Commun
481:146-152
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Arabidopsis thaliana SNC1 TIR domain.
|
|
K.G.Hyun,
Y.Lee,
J.Yoon,
H.Yi,
J.J.Song.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Plant immune response is initiated by Resistance proteins (R proteins).
Toll/interleukin-1 receptor (TIR) domain in R proteins, which is responsible for
the dimerization but has limited conservation in their primary structures.
Suppressor of npr1-1, constitutive 1 (SNC1), a TIR-containing R protein, is
involved in autoimmunity of plant, but the binding partner of SNC1 via the TIR
domain and its specific cognate effector protein remain elusive. Here, we
present the crystal structure of the TIR domain of Arabidopsis thaliana SNC1
(AtSNC1-TIR). The structure shows that AtSNC1-TIR domain is similar to those of
other plant TIR domains including AtTIR, L6 and RPS4. Structural and sequence
analysis on AtSNC1-TIR revealed that almost all conserved amino acids are
located in the core of the structure, while the amino acids on the surface are
highly variable, implicating that each TIR domain utilizes the variable surface
for interacting its binding partner. In addition, the interaction between
AtSNC1-TIR proteins in the crystal suggests two possible dimerization modes of
AtSNC1-TIR domain. This study provides structural platform to investigate
AtSNC1-TIR mediated signaling pathway of plant immune responses.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

