 |
PDBsum entry 5b4k

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase inhibitor
|
PDB id
|
|

|

|
5b4k
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.17
- 3',5'-cyclic-nucleotide phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate + H+
|
 |
 |
 |
 |
 |
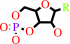 nucleoside 3',5'-cyclic phosphate
nucleoside 3',5'-cyclic phosphate
|
+
|
H2O
|
=
|
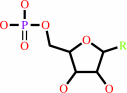 nucleoside 5'-phosphate
nucleoside 5'-phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Bioorg Med Chem Lett
24:3447-3455
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Design and synthesis of potent and selective pyridazin-4(1H)-one-based PDE10A inhibitors interacting with Tyr683 in the PDE10A selectivity pocket.
|
|
M.Yoshikawa,
T.Hitaka,
T.Hasui,
M.Fushimi,
J.Kunitomo,
H.Kokubo,
H.Oki,
K.Nakashima,
T.Taniguchi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Utilizing structure-based drug design techniques, we designed and synthesized
phosphodiesterase 10A (PDE10A) inhibitors based on pyridazin-4(1H)-one. These
compounds can interact with Tyr683 in the PDE10A selectivity pocket.
Pyridazin-4(1H)-one derivative 1 was linked with a benzimidazole group through
an alkyl spacer to interact with the OH of Tyr683 and fill the PDE10A
selectivity pocket. After optimizing the linker length, we identified
1-(cyclopropylmethyl)-5-[3-(1-methyl-1H-benzimidazol-2-yl)propoxy]-3-(1-phenyl-1H-pyrazol-5-yl)pyridazin-4(1H)-one
(16f) as having highly potent PDE10A inhibitory activity (IC50=0.76nM) and
perfect selectivity against other PDEs (>13,000-fold, IC50=>10,000nM). The
crystal structure of 16f bound to PDE10A revealed that the benzimidazole moiety
was located deep within the PDE10A selectivity pocket and interacted with
Tyr683. Additionally, a bidentate interaction existed between the
5-alkoxypyridazin-4(1H)-one moiety and the conserved Gln716 present in all PDEs.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

