 |
PDBsum entry 5hft

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 305 a.a.
305 a.a.
|
 |

|

|

|

|

|

|

|
 149 a.a.
149 a.a.
|
 |

|

|

|

|

|

|

|
 154 a.a.
154 a.a.
|
 |

|

|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of hpxw
|
|
Structure:
|
 |
Gamma-glutamyltranspeptidase. Chain: a, c. Fragment: residues 1-341. Engineered: yes. Gamma-glutamyltranspeptidase. Chain: b, d. Fragment: residues 342-528. Engineered: yes
|
|
Source:
|
 |
Klebsiella pneumoniae subsp. Pneumoniae. Organism_taxid: 272620. Strain: atcc 700721 / mgh 78578. Gene: kpn_01768. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.65Å
|
R-factor:
|
0.257
|
R-free:
|
0.298
|
|
|
Authors:
|
 |
S.E.Ealick,K.A.Hicks
|
|
Key ref:
|
 |
K.A.Hicks
and
S.E.Ealick
(2016).
Biochemical and structural characterization of Klebsiella pneumoniae oxamate amidohydrolase in the uric acid degradation pathway.
Acta Crystallogr D Struct Biol,
72,
808-816.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
07-Jan-16
|
Release date:
|
29-Jun-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A6T9C8
(HPXW_KLEP7) -
Oxamate amidohydrolase proenzyme from Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
528 a.a.
305 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, C, B, D:
E.C.3.5.1.126
- oxamate amidohydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxamate + H2O = oxalate + NH4+
|
 |
 |
 |
 |
 |
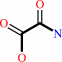 oxamate
oxamate
|
+
|
H2O
|
=
|
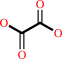 oxalate
oxalate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Struct Biol
72:808-816
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Biochemical and structural characterization of Klebsiella pneumoniae oxamate amidohydrolase in the uric acid degradation pathway.
|
|
K.A.Hicks,
S.E.Ealick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
HpxW from the ubiquitous pathogen Klebsiella pneumoniae is involved in a novel
uric acid degradation pathway downstream from the formation of oxalurate.
Specifically, HpxW is an oxamate amidohydrolase which catalyzes the conversion
of oxamate to oxalate and is a member of the Ntn-hydrolase superfamily. HpxW is
autoprocessed from an inactive precursor to form a heterodimer, resulting in a
35.5 kDa α subunit and a 20 kDa β subunit. Here, the structure of HpxW is
presented and the substrate complex is modeled. In addition, the steady-state
kinetics of this enzyme and two active-site variants were characterized. These
structural and biochemical studies provide further insight into this class of
enzymes and allow a mechanism for catalysis consistent with other members of the
Ntn-hydrolase superfamily to be proposed.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| |

