 |
PDBsum entry 4qj4

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Signaling protein/hydrolase
|
PDB id
|
|

|

|
4qj4
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain B:
E.C.3.1.4.11
- phosphoinositide phospholipase C.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Phosphate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + H2O = 1D-myo-inositol 1,4,5-trisphosphate + a 1,2-diacyl-sn-glycerol + H+
|
 |
 |
 |
 |
 |
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate)
|
+
|
H2O
|
=
|
1D-myo-inositol 1,4,5-trisphosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
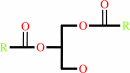 1,2-diacyl-sn-glycerol
1,2-diacyl-sn-glycerol
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
22:1844-1854
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Molecular mechanisms of phospholipase C β3 autoinhibition.
|
|
A.M.Lyon,
J.A.Begley,
T.D.Manett,
J.J.Tesmer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phospholipase C β (PLCβ) enzymes are dramatically activated by heterotrimeric
G proteins. Central to this response is the robust autoinhibition of PLCβ by
the X-Y linker region within its catalytic core and by the Hα2' helix in the
C-terminal extension of the enzyme. The molecular mechanism of each and their
mutual dependence are poorly understood. Herein, it is shown that distinct
regions within the X-Y linker have specific roles in regulating activity. Most
important,an acidic stretch within the linker stabilizes a lid that occludes the
active site, consistent with crystal structures of variants lacking this region.
Inhibition by the Hα2' helix is independent of the X-Y linker and likely
regulates activity by limiting membrane interaction of the catalytic core. Full
activation of PLCβ thus requires multiple independent molecular events induced
by membrane association of the catalytic core and by the binding of regulatory
proteins.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

