 |
PDBsum entry 4e75
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of lpxd from acinetobacter baumannii at 2.85a resolution (p21 form)
|
|
Structure:
|
 |
Udp-3-o-acylglucosamine n-acyltransferase. Chain: a, b, c, d, e, f. Engineered: yes
|
|
Source:
|
 |
Acinetobacter baumannii. Organism_taxid: 470. Gene: lpxd, absdf1688. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_cell_line: bl21 (de3)
|
|
Resolution:
|
 |
|
2.85Å
|
R-factor:
|
0.237
|
R-free:
|
0.280
|
|
|
Authors:
|
 |
J.Badger,B.Chie-Leon,C.Logan,V.Sridhar,B.Sankaran,P.H.Zwart, V.Nienaber
|
|
Key ref:
|
 |
J.Badger
et al.
(2013).
Structure determination of LpxD from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
Acta Crystallogr Sect F Struct Biol Cryst Commun,
69,
6-9.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
16-Mar-12
|
Release date:
|
16-Jan-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
B0VMV2
(LPXD_ACIBS) -
UDP-3-O-acylglucosamine N-acyltransferase from Acinetobacter baumannii (strain SDF)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
356 a.a.
337 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.191
- UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a UDP-3-O-[(3R)-3-hydroxyacyl]-alpha-D-glucosamine + a (3R)-hydroxyacyl- [ACP] = a UDP-2-N,3-O-bis[(3R)-3-hydroxyacyl]-alpha-D-glucosamine + holo- [ACP] + H+
|
 |
 |
 |
 |
 |
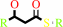 (3R)-3-hydroxyacyl-[acyl-carrier-protein]
(3R)-3-hydroxyacyl-[acyl-carrier-protein]
|
+
|
UDP-3-O-((3R)- hydroxyacyl)-alpha-D-glucosamine
|
=
|
UDP-2-N,3-O-bis((3R)-3-hydroxyacyl)- alpha-D-glucosamine
|
+
|
 holo-[acyl-carrier-protein]
holo-[acyl-carrier-protein]
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr Sect F Struct Biol Cryst Commun
69:6-9
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure determination of LpxD from the lipopolysaccharide-synthesis pathway of Acinetobacter baumannii.
|
|
J.Badger,
B.Chie-Leon,
C.Logan,
V.Sridhar,
B.Sankaran,
P.H.Zwart,
V.Nienaber.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Acinetobacter baumannii is a Gram-negative bacterium that is resistant to many
currently available antibiotics. The protein LpxD is a component of the
biosynthetic pathway for lipopolysaccharides in the outer membrane of this
bacterium and is a potential target for new antibacterial agents. This paper
describes the structure determination of apo forms of LpxD in space groups P2(1)
and P4(3)22. These crystals contained six and three copies of the protein
molecule in the asymmetric unit and diffracted to 2.8 and 2.7 Å resolution,
respectively. A comparison of the multiple protein copies in the asymmetric
units of these crystals reveals a common protein conformation and a conformation
in which the relative orientation between the two major domains in the protein
is altered.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

