 |
PDBsum entry 4ajd
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Identification and structural characterization of pde10 fragment inhibitors
|
|
Structure:
|
 |
Camp and camp-inhibited cgmp 3', 5'-cyclic phosphodiesterase 10a, pde10. Chain: a, d. Fragment: catalytic domain, residues 439-764. Synonym: phosphodiesterase 10a. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.190
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
P.Johansson,J.S.Albert,L.Spadola,T.Akerud,E.Back,P.Hillertz, R.Horsefeld,C.Scott,N.Spear,G.Tian,A.Tigerstrom,D.Aharony, S.Geschwindner
|
|
Key ref:
|
 |
P.Johansson
et al.
Identification and structural characterization of pde fragment inhibitors.
To be published,
.
|
 |
|
Date:
|
 |
|
16-Feb-12
|
Release date:
|
06-Mar-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9Y233
(PDE10_HUMAN) -
cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1055 a.a.
324 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.17
- 3',5'-cyclic-nucleotide phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate + H+
|
 |
 |
 |
 |
 |
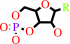 nucleoside 3',5'-cyclic phosphate
nucleoside 3',5'-cyclic phosphate
|
+
|
H2O
|
=
|
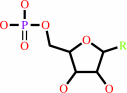 nucleoside 5'-phosphate
nucleoside 5'-phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

