 |
PDBsum entry 3ohm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Signaling protein / hydrolase
|
PDB id
|
|

|

|
3ohm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain B:
E.C.3.1.4.11
- phosphoinositide phospholipase C.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Phosphate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + H2O = 1D-myo-inositol 1,4,5-trisphosphate + a 1,2-diacyl-sn-glycerol + H+
|
 |
 |
 |
 |
 |
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate)
|
+
|
H2O
|
=
|
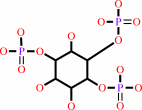 1D-myo-inositol 1,4,5-trisphosphate
1D-myo-inositol 1,4,5-trisphosphate
|
+
|
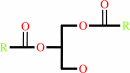 1,2-diacyl-sn-glycerol
1,2-diacyl-sn-glycerol
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Science
330:974-980
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Kinetic scaffolding mediated by a phospholipase C-beta and Gq signaling complex.
|
|
G.L.Waldo,
T.K.Ricks,
S.N.Hicks,
M.L.Cheever,
T.Kawano,
K.Tsuboi,
X.Wang,
C.Montell,
T.Kozasa,
J.Sondek,
T.K.Harden.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.M.Lyon,
V.M.Tesmer,
V.D.Dhamsania,
D.M.Thal,
J.Gutierrez,
S.Chowdhury,
K.C.Suddala,
J.K.Northup,
and
J.J.Tesmer
(2011).
An autoinhibitory helix in the C-terminal region of phospholipase C-β mediates Gαq activation.
|
| |
Nat Struct Mol Biol,
18,
999.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.H.Moolenaar,
and
A.Perrakis
(2011).
Insights into autotaxin: how to produce and present a lipid mediator.
|
| |
Nat Rev Mol Cell Biol,
12,
674-679.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

