 |
PDBsum entry 2exr

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2exr
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
X-ray structure of cytokinin oxidase/dehydrogenase (ckx) from arabidopsis thaliana at5g21482
|
|
Structure:
|
 |
Cytokinin dehydrogenase 7. Chain: a. Synonym: cytokinin oxidase 7, cko7, atckx7, atckx5. Engineered: yes
|
|
Source:
|
 |
Arabidopsis thaliana. Thale cress. Organism_taxid: 3702. Gene: ckx7, ckx5. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.173
|
R-free:
|
0.214
|
|
|
Authors:
|
 |
G.E.Wesenberg,G.N.Phillips Jr.,B.W.Han,E.Bitto,C.A.Bingman,E.Bae, Center For Eukaryotic Structural Genomics (Cesg)
|
Key ref:

|
 |
E.Bae
et al.
(2008).
Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase.
Proteins,
70,
303-306.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
08-Nov-05
|
Release date:
|
29-Nov-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9FUJ1
(CKX7_ARATH) -
Cytokinin dehydrogenase 7 from Arabidopsis thaliana
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
524 a.a.
491 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.5.99.12
- cytokinin dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-dimethylallyladenine + A + H2O = 3-methyl-2-butenal + adenine + AH2
|
 |
 |
 |
 |
 |
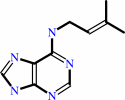 N(6)-dimethylallyladenine
N(6)-dimethylallyladenine
|
+
|
|
+
|
H2O
|
=
|
3-methyl-2-butenal
|
+
|
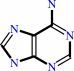 adenine
adenine
|
+
|
AH2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
70:303-306
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Arabidopsis thaliana cytokinin dehydrogenase.
|
|
E.Bae,
C.A.Bingman,
E.Bitto,
D.J.Aceti,
G.N.Phillips.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1. Crystal structure of AtCKX7. (A) Ribbon diagram of
AtCKX7. The FAD-binding and the substrate-binding domains are
shown in cyan and red, respectively. The FAD molecule is
represented as a stick model in yellow. (B) Covalent bond
between His96 and the isoalloxazine ring of the FAD molecule.
The FAD is shown in yellow and the His residue is colored by
atom types (carbon: green; oxygen: red; nitrogen: blue). The
2F[obs] - F[calc] map is contoured at 1.5  . .
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2008,
70,
303-306)
copyright 2008.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Werner,
and
T.Schmülling
(2009).
Cytokinin action in plant development.
|
| |
Curr Opin Plant Biol,
12,
527-538.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

