 |
PDBsum entry 2drb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/RNA
|
PDB id
|
|

|

|
2drb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.72
- Cca tRNA nucleotidyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a tRNA precursor + 2 CTP + ATP = a tRNA with a 3' CCA end + 3 diphosphate
|
 |
 |
 |
 |
 |
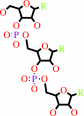 tRNA precursor
tRNA precursor
|
+
|
 2
×
CTP
2
×
CTP
|
+
|
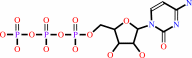
 ATP
ATP
|
=
|
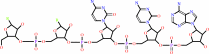 tRNA with a 3' CCA end
tRNA with a 3' CCA end
|
+
|
 3
×
diphosphate
3
×
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
443:956-960
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Complete crystallographic analysis of the dynamics of CCA sequence addition.
|
|
K.Tomita,
R.Ishitani,
S.Fukai,
O.Nureki.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
CCA-adding polymerase matures the essential 3'-CCA terminus of transfer RNA
without any nucleic-acid template. However, it remains unclear how the correct
nucleotide triphosphate is selected in each reaction step and how the
polymerization is driven by the protein and RNA dynamics. Here we present
complete sequential snapshots of six complex structures of CCA-adding enzyme and
four distinct RNA substrates with and without CTP (cytosine triphosphate) or ATP
(adenosine triphosphate). The CCA-lacking RNA stem extends by one base pair to
force the discriminator nucleoside into the active-site pocket, and then tracks
back after incorporation of the first cytosine monophosphate (CMP).
Accommodation of the second CTP clamps the catalytic cleft, inducing a
reorientation of the turn, which flips C74 to allow CMP to be accepted. In
contrast, after the second CMP is added, the polymerase and RNA primer are
locked in the closed state, which directs the subsequent A addition. Between the
CTP- and ATP-binding stages, the side-chain conformation of Arg 224 changes
markedly; this is controlled by the global motion of the enzyme and position of
the primer terminus, and is likely to achieve the CTP/ATP discrimination,
depending on the polymerization stage. Throughout the CCA-adding reaction, the
enzyme tail domain firmly anchors the TPsiC-loop of the tRNA, which ensures
accurate polymerization and termination.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2: Expansion and contraction of the primer RNA helix at
the mini-D stage. a, Extended mini-helix structure at the
mini-D stage. The simulated annealed omit maps (contoured at 3.5
 )
for the indicated nucleosides are shown. b, Back-tracked
mini-helix structure at the mini-DC. Other stages have the same
standard helix structure. )
for the indicated nucleosides are shown. b, Back-tracked
mini-helix structure at the mini-DC. Other stages have the same
standard helix structure.
|
 |
Figure 3.
Figure 3: Active-site structure at each reaction stage. a,
Mini-D stage (the simulated annealed omit maps contoured at 4
 for
G1 and A73 are shown). b, Mini-DC stage. c, Mini-DC + CTP stage.
d, Mini-DCC stage. e, Mini-DCC + ATP stage. f, Mini-DCCA stage.
g, Mature tRNA dissociation stage (PDB ID: 1SZ1)^14. The
acceptor stem expansion at the first CCA-adding step can easily
be seen by the fact that Asp 291 recognizes the 2'-OH group of
C72 in the mini-D stage, but A73 in the other stages. The
catalytic triad comprises Glu 59, Asp 61 and Asp 110. The
hydrogen bonds are represented by dotted lines. for
G1 and A73 are shown). b, Mini-DC stage. c, Mini-DC + CTP stage.
d, Mini-DCC stage. e, Mini-DCC + ATP stage. f, Mini-DCCA stage.
g, Mature tRNA dissociation stage (PDB ID: 1SZ1)^14. The
acceptor stem expansion at the first CCA-adding step can easily
be seen by the fact that Asp 291 recognizes the 2'-OH group of
C72 in the mini-D stage, but A73 in the other stages. The
catalytic triad comprises Glu 59, Asp 61 and Asp 110. The
hydrogen bonds are represented by dotted lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nature
(2006,
443,
956-960)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.L.Gleghorn,
E.K.Davydova,
R.Basu,
L.B.Rothman-Denes,
and
K.S.Murakami
(2011).
X-ray crystal structures elucidate the nucleotidyl transfer reaction of transcript initiation using two nucleotides.
|
| |
Proc Natl Acad Sci U S A,
108,
3566-3571.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Bai,
S.K.Srivastava,
J.H.Chang,
J.L.Manley,
and
L.Tong
(2011).
Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase.
|
| |
Mol Cell,
41,
311-320.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Hoffmeier,
H.Betat,
A.Bluschke,
R.Günther,
S.Junghanns,
H.J.Hofmann,
and
M.Mörl
(2010).
Unusual evolution of a catalytic core element in CCA-adding enzymes.
|
| |
Nucleic Acids Res,
38,
4436-4447.
|
 |
|
|
|
|
 |
B.Pan,
Y.Xiong,
and
T.A.Steitz
(2010).
How the CCA-adding enzyme selects adenine over cytosine at position 76 of tRNA.
|
| |
Science,
330,
937-940.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Betat,
C.Rammelt,
and
M.Mörl
(2010).
tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization.
|
| |
Cell Mol Life Sci,
67,
1447-1463.
|
 |
|
|
|
|
 |
I.U.Heinemann,
D.Söll,
and
L.Randau
(2010).
Transfer RNA processing in archaea: unusual pathways and enzymes.
|
| |
FEBS Lett,
584,
303-309.
|
 |
|
|
|
|
 |
M.Blaise,
M.Bailly,
M.Frechin,
M.A.Behrens,
F.Fischer,
C.L.Oliveira,
H.D.Becker,
J.S.Pedersen,
S.Thirup,
and
D.Kern
(2010).
Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation.
|
| |
EMBO J,
29,
3118-3129.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Giegé,
and
C.Sauter
(2010).
Biocrystallography: past, present, future.
|
| |
HFSP J,
4,
109-121.
|
 |
|
|
|
|
 |
S.Hamill,
S.L.Wolin,
and
K.M.Reinisch
(2010).
Structure and function of the polymerase core of TRAMP, a RNA surveillance complex.
|
| |
Proc Natl Acad Sci U S A,
107,
15045-15050.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.M.Hou
(2010).
CCA addition to tRNA: implications for tRNA quality control.
|
| |
IUBMB Life,
62,
251-260.
|
 |
|
|
|
|
 |
S.Kim,
C.Liu,
K.Halkidis,
H.B.Gamper,
and
Y.M.Hou
(2009).
Distinct kinetic determinants for the stepwise CCA addition to tRNA.
|
| |
RNA,
15,
1827-1836.
|
 |
|
|
|
|
 |
Y.Toh,
D.Takeshita,
T.Numata,
S.Fukai,
O.Nureki,
and
K.Tomita
(2009).
Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme.
|
| |
EMBO J,
28,
3353-3365.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.D.Cho,
V.D.Sood,
D.Baker,
and
A.M.Weiner
(2008).
On the role of a conserved, potentially helix-breaking residue in the tRNA-binding alpha-helix of archaeal CCA-adding enzymes.
|
| |
RNA,
14,
1284-1289.
|
 |
|
|
|
|
 |
M.Dupasquier,
S.Kim,
K.Halkidis,
H.Gamper,
and
Y.M.Hou
(2008).
tRNA integrity is a prerequisite for rapid CCA addition: implication for quality control.
|
| |
J Mol Biol,
379,
579-588.
|
 |
|
|
|
|
 |
R.Giegé
(2008).
Toward a more complete view of tRNA biology.
|
| |
Nat Struct Mol Biol,
15,
1007-1014.
|
 |
|
|
|
|
 |
X.Shan,
T.A.Russell,
S.M.Paul,
D.B.Kushner,
and
P.B.Joyce
(2008).
Characterization of a temperature-sensitive mutation that impairs the function of yeast tRNA nucleotidyltransferase.
|
| |
Yeast,
25,
219-233.
|
 |
|
|
|
|
 |
Y.Toh,
T.Numata,
K.Watanabe,
D.Takeshita,
O.Nureki,
and
K.Tomita
(2008).
Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme.
|
| |
EMBO J,
27,
1944-1952.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Martin,
and
W.Keller
(2007).
RNA-specific ribonucleotidyl transferases.
|
| |
RNA,
13,
1834-1849.
|
 |
|
|
|
|
 |
H.Li
(2007).
Complexes of tRNA and maturation enzymes: shaping up for translation.
|
| |
Curr Opin Struct Biol,
17,
293-301.
|
 |
|
|
|
|
 |
S.Muller,
J.B.Fourmann,
C.Loegler,
B.Charpentier,
and
C.Branlant
(2007).
Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:Psi55-synthase and RNA-guided RNA:Psi-synthase activities.
|
| |
Nucleic Acids Res,
35,
5610-5624.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

