 |
PDBsum entry 1qnv

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.24
- porphobilinogen synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Porphyrin Biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 5-aminolevulinate = porphobilinogen + 2 H2O + H+
|
 |
 |
 |
 |
 |
 2
×
5-aminolevulinate
2
×
5-aminolevulinate
|
=
|
 porphobilinogen
porphobilinogen
|
+
|
2
×
H2O
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
56:421-430
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
MAD analyses of yeast 5-aminolaevulinate dehydratase: their use in structure determination and in defining the metal-binding sites.
|
|
P.T.Erskine,
E.M.Duke,
I.J.Tickle,
N.M.Senior,
M.J.Warren,
J.B.Cooper.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
MAD experiments attempting to solve the structure of 5--aminolaevulinic acid
dehydratase using Zn and Pb edges are described. The data obtained proved
insufficient for a complete structure solution but were invaluable in subsequent
identification of metal-binding sites using anomalous difference Fourier
analyses once the structure of the enzyme had been solved. These sites include
the highly inhibitory substitution of an enzymic cofactor Zn(2+) ion by Pb(2+)
ions, which represents a major contribution towards understanding the molecular
basis of lead poisoning. The MAD data collected at the Pb edge were also used
with isomorphous replacement data from the same Pb co-crystal and a Hg
co-crystal to provide the first delineation of the enzyme's quaternary
structure. In this MADIR analysis, the Hg co-crystal data were treated as native
data. Anomalous difference Fouriers were again used, revealing that Hg(2+) had
substituted for the same Zn(2+) cofactor ion as had Pb(2+), a finding of
fundamental importance for the understanding of mercury poisoning. In addition,
Pt(2+) ions were found to bind at the same place in the structure. The refined
structures of the Pb- and the Hg-complexed enzymes are presented at 2.5 and 3.0
A resolution, respectively.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4 S. cerevisiae ALAD Pb edge (PX 9.5 MAD 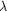 [1]
data) anomalous difference Patterson at 3.0 Å resolution, u = ½
Harker section. The site marked by a cross was tried in a
superposition function in VECSUM but failed to bring back other
sites. It produced poor phasing statistics in MLPHARE. From the
isomorphous replacement processing of the PX 9.6 data from this
crystal against the isomorphous Hg co-crystal the site
corresponds to one of the Pb sites. The major axial peak at the
right corresponds to peaks in that processing found to have
arisen from cross vectors between the two lead sites. The map is
contoured at 94% r.m.s. [1]
data) anomalous difference Patterson at 3.0 Å resolution, u = ½
Harker section. The site marked by a cross was tried in a
superposition function in VECSUM but failed to bring back other
sites. It produced poor phasing statistics in MLPHARE. From the
isomorphous replacement processing of the PX 9.6 data from this
crystal against the isomorphous Hg co-crystal the site
corresponds to one of the Pb sites. The major axial peak at the
right corresponds to peaks in that processing found to have
arisen from cross vectors between the two lead sites. The map is
contoured at 94% r.m.s.
|
 |
Figure 6.
Figure 6 A MADIR approach with data from the S. cerevisiae ALAD
lead acetate co-crystal allowed the calculation of an
electron-density map at 3.0 Å resolution in which there was a
clear solvent boundary. The map was extended to show the overall
organization of the octamer. Here, four subunits of the ALAD
octamer can be seen disposed around the crystallographic
fourfold axis.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2000,
56,
421-430)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
O.Iranzo,
T.Jakusch,
K.H.Lee,
L.Hemmingsen,
and
V.L.Pecoraro
(2009).
The correlation of 113Cd NMR and 111mCd PAC spectroscopies provides a powerful approach for the characterization of the structure of Cd(II)-substituted Zn(II) proteins.
|
| |
Chemistry,
15,
3761-3772.
|
 |
|
|
|
|
 |
M.Kirberger,
and
J.J.Yang
(2008).
Structural differences between Pb2+- and Ca2+-binding sites in proteins: implications with respect to toxicity.
|
| |
J Inorg Biochem,
102,
1901-1909.
|
 |
|
|
|
|
 |
D.S.Touw,
C.E.Nordman,
J.A.Stuckey,
and
V.L.Pecoraro
(2007).
Identifying important structural characteristics of arsenic resistance proteins by using designed three-stranded coiled coils.
|
| |
Proc Natl Acad Sci U S A,
104,
11969-11974.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Matzapetakis,
D.Ghosh,
T.C.Weng,
J.E.Penner-Hahn,
and
V.L.Pecoraro
(2006).
Peptidic models for the binding of Pb(II), Bi(III) and Cd(II) to mononuclear thiolate binding sites.
|
| |
J Biol Inorg Chem,
11,
876-890.
|
 |
|
|
|
|
 |
N.Sawada,
N.Nagahara,
T.Sakai,
Y.Nakajima,
M.Minami,
and
T.Kawada
(2005).
The activation mechanism of human porphobilinogen synthase by 2-mercaptoethanol: intrasubunit transfer of a reserve zinc ion and coordination with three cysteines in the active center.
|
| |
J Biol Inorg Chem,
10,
199-207.
|
 |
|
|
|
|
 |
L.Kundrat,
J.Martins,
L.Stith,
R.L.Dunbrack,
and
E.K.Jaffe
(2003).
A structural basis for half-of-the-sites metal binding revealed in Drosophila melanogaster porphobilinogen synthase.
|
| |
J Biol Chem,
278,
31325-31330.
|
 |
|
|
|
|
 |
S.Breinig,
J.Kervinen,
L.Stith,
A.S.Wasson,
R.Fairman,
A.Wlodawer,
A.Zdanov,
and
E.K.Jaffe
(2003).
Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase.
|
| |
Nat Struct Biol,
10,
757-763.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Kim,
and
D.M.Ogrydziak
(2000).
Current awareness on yeast.
|
| |
Yeast,
16,
1253-1260.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

