 |
PDBsum entry 1kho

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.3
- phospholipase C.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phosphocholine + H2O = phosphocholine + a 1,2- diacyl-sn-glycerol + H+
|
 |
 |
 |
 |
 |
 1,2-diacyl-sn-glycero-3-phosphocholine
1,2-diacyl-sn-glycero-3-phosphocholine
|
+
|
H2O
|
=
|
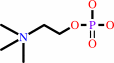 phosphocholine
phosphocholine
|
+
|
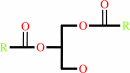 1,2- diacyl-sn-glycerol
1,2- diacyl-sn-glycerol
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:6253-6262
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
The first strain of Clostridium perfringens isolated from an avian source has an alpha-toxin with divergent structural and kinetic properties.
|
|
N.Justin,
N.Walker,
H.L.Bullifent,
G.Songer,
D.M.Bueschel,
H.Jost,
C.Naylor,
J.Miller,
D.S.Moss,
R.W.Titball,
A.K.Basak.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Clostridium perfringens alpha-toxin is a 370-residue, zinc-dependent,
phospholipase C that is the key virulence determinant in gas gangrene. It is
also implicated in the pathogenesis of sudden death syndrome in young animals
and necrotic enteritis in chickens. Previously characterized alpha-toxins from
different strains of C. perfringens are almost identical in sequence and
biochemical properties. We describe the cloning, nucleotide sequencing,
expression, characterization, and crystal structure of alpha-toxin from an avian
strain, SWan C. perfringens (SWCP), which has a large degree of sequence
variation and altered substrate specificity compared to these strains. The
structure of alpha-toxin from strain CER89L43 has been previously reported in
open (active site accessible to substrate) and closed (active site obscured by
loop movements) conformations. The SWCP structure is in an open-form
conformation, with three zinc ions in the active site. This is the first example
of an open form of alpha-toxin crystallizing without the addition of divalent
cations to the crystallization buffer, indicating that the protein can retain
three zinc ions bound in the active site. The topology of the calcium binding
site formed by residues 269, 271, 336, and 337, which is essential for membrane
binding, is significantly altered in comparison with both the open and closed
alpha-toxin structures. We are able to relate these structural changes to the
different substrate specificity and membrane binding properties of this
divergent alpha-toxin. This will provide essential information when developing
an effective vaccine that will protect against C. perfringens infection in a
wide range of domestic livestock.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.G.Vachieri,
G.C.Clark,
A.Alape-Girón,
M.Flores-Díaz,
N.Justin,
C.E.Naylor,
R.W.Titball,
and
A.K.Basak
(2010).
Comparison of a nontoxic variant of Clostridium perfringens α-toxin with the toxic wild-type strain.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
1067-1074.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.R.Popoff,
and
P.Bouvet
(2009).
Clostridial toxins.
|
| |
Future Microbiol,
4,
1021-1064.
|
 |
|
|
|
|
 |
D.R.Thompson,
V.R.Parreira,
R.R.Kulkarni,
and
J.F.Prescott
(2006).
Live attenuated vaccine-based control of necrotic enteritis of broiler chickens.
|
| |
Vet Microbiol,
113,
25-34.
|
 |
|
|
|
|
 |
S.A.Sheedy,
A.B.Ingham,
J.I.Rood,
and
R.J.Moore
(2004).
Highly conserved alpha-toxin sequences of avian isolates of Clostridium perfringens.
|
| |
J Clin Microbiol,
42,
1345-1347.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

