
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chain B:
E.C.2.3.1.48
- histone acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-lysyl-[protein] + acetyl-CoA = N6-acetyl-L-lysyl-[protein] + CoA + H+
|
 |
 |
 |
 |
 |
L-lysyl-[protein]
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
N(6)-acetyl-L-lysyl-[protein]
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain B:
E.C.2.3.1.57
- diamine N-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an alkane-alpha,omega-diamine + acetyl-CoA = an N-acetylalkane- alpha,omega-diamine + CoA + H+
|
 |
 |
 |
 |
 |
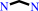 alkane-alpha,omega-diamine
alkane-alpha,omega-diamine
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
N-acetylalkane- alpha,omega-diamine
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Mol Cell
9:575-586
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain.
|
|
S.Mujtaba,
Y.He,
L.Zeng,
A.Farooq,
J.E.Carlson,
M.Ott,
E.Verdin,
M.M.Zhou.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The human immunodeficiency virus type 1 (HIV-1) trans-activator protein Tat
stimulates transcription of the integrated HIV-1 genome and promotes viral
replication in infected cells. Tat transactivation activity is dependent on
lysine acetylation and its association with nuclear histone acetyltransferases
p300/CBP (CREB binding protein) and p300/CBP-associated factor (PCAF). Here, we
show that the bromodomain of PCAF binds specifically to HIV-1 Tat acetylated at
lysine 50 and that this interaction competes effectively against HIV-1 TAR RNA
binding to the lysine-acetylated Tat. The three-dimensional solution structure
of the PCAF bromodomain in complex with a lysine 50-acetylated Tat peptide
together with biochemical analyses provides the structural basis for the
specificity of this molecular recognition and reveals insights into the
differences in ligand selectivity of bromodomains.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. NMR Structure of the PCAF Bromodomain/Tat AcK50
Peptide Complex(A) Stereoview of the backbone atoms (N, Cα, and
C′) of 25 superimposed NMR-derived structures of the PCAF
bromodomain (black) (showing residues 719–830) in complex with
the Tat AcK50 peptide (green) (showing residues 46–55). Note
that amino acid residues in the Tat peptide are described either
according to their relative positions with respect to the
acetyl-lysine in the sequence or for clarity numbered by their
specific positions in the protein sequence of Tat.(B) Ribbons
(Carson, 1991) representation of the average minimized NMR
structure of the PCAF bromodomain/Tat peptide complex.(C)
Stereoview of the Tat binding site in the bromodomain showing
side chains of the protein (green) and peptide (blue) residues
that are directly involved intermolecular interactions.(D)
Stereoview of superimposition of the free (green) and
ligand-bound (blue) structures of PCAF bromodomain showing side
chain conformation of the residues in the Tat peptide binding
site. The residues of the Tat peptide are colored in orange.(E)
Surface representation of the Tat binding site of the
bromodomain in ligand-bound (left) and free form (right).
Protein residues important in ligand recognition are colored
with the same color scheme in both structures. Residues
indicated by an asterisk are almost completely buried in the
free form structure.
|
 |
Figure 4.
Figure 4. PCAF Bromodomain Competing against TAR RNA for
Binding to Tat AcK50 Peptide(A) Superimposition of a selected
region of 2D ^1H-^15N HSQC spectra of a ^13C/^15N-labeled PCAF
bromodomain protein in the free form (black), in the presence of
Tat AcK50 peptide (molar ratio of 1:1.2) (red), and in the
presence of Tat AcK50 peptide and TAR RNA (molar ratio of
1:1.2:1 for protein:peptide:RNA) (blue). The spectra show
chemical shift changes of the backbone amide resonances of
protein residues due to peptide binding.(B) Superimposition of
2D ^1H-^13C HSQC spectra of the PCAF bromodomain collected under
the same conditions as described in (A). The NMR spectra exhibit
chemical shift changes of the side chain methyl group resonances
of protein residues due to peptide binding. The same
color-coding scheme was used as in (A). Arrows indicate chemical
shift changes of protein NMR resonances from the free form
(black) to the Tat AcK50 peptide-bound form (red). Note that
only the bromodomain residues (i.e., A757 and V752) that
directly interact with the Tat peptide, as shown in the
three-dimensional structure, exhibited major chemical shift
changes upon peptide binding or in competing against TAR RNA for
binding to the Tat peptide. More importantly, addition of TAR
RNA causes only small shifts of the protein signals from the Tat
peptide-bound position toward the free form position, suggesting
that the PCAF bromodomain competes effectively against TAR RNA
for binding to the lysine-acetylated Tat peptide. We observed by
NMR no significant nonspecific interactions between the protein
and TAR RNA under these conditions.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Mol Cell
(2002,
9,
575-586)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Patel,
R.R.Pathak,
and
S.Mujtaba
(2011).
The biology of lysine acetylation integrates transcriptional programming and metabolism.
|
| |
Nutr Metab (Lond),
8,
12.
|
 |
|
|
|
|
 |
M.K.Johri,
R.Mishra,
C.Chhatbar,
S.K.Unni,
and
S.K.Singh
(2011).
Tits and bits of HIV Tat protein.
|
| |
Expert Opin Biol Ther,
11,
269-283.
|
 |
|
|
|
|
 |
S.Sengupta,
A.K.Mantha,
S.Mitra,
and
K.K.Bhakat
(2011).
Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1.
|
| |
Oncogene,
30,
482-493.
|
 |
|
|
|
|
 |
J.I.MacPherson,
J.E.Dickerson,
J.W.Pinney,
and
D.L.Robertson
(2010).
Patterns of HIV-1 protein interaction identify perturbed host-cellular subsystems.
|
| |
PLoS Comput Biol,
6,
e1000863.
|
 |
|
|
|
|
 |
K.L.Yap,
and
M.M.Zhou
(2010).
Keeping it in the family: diverse histone recognition by conserved structural folds.
|
| |
Crit Rev Biochem Mol Biol,
45,
488-505.
|
 |
|
|
|
|
 |
Q.Zhang,
S.Chakravarty,
D.Ghersi,
L.Zeng,
A.N.Plotnikov,
R.Sanchez,
and
M.M.Zhou
(2010).
Biochemical profiling of histone binding selectivity of the yeast bromodomain family.
|
| |
PLoS One,
5,
e8903.
|
 |
|
|
|
|
 |
R.Easley,
R.Van Duyne,
W.Coley,
I.Guendel,
S.Dadgar,
K.Kehn-Hall,
and
F.Kashanchi
(2010).
Chromatin dynamics associated with HIV-1 Tat-activated transcription.
|
| |
Biochim Biophys Acta,
1799,
275-285.
|
 |
|
|
|
|
 |
S.Pagans,
S.E.Kauder,
K.Kaehlcke,
N.Sakane,
S.Schroeder,
W.Dormeyer,
R.C.Trievel,
E.Verdin,
M.Schnolzer,
and
M.Ott
(2010).
The Cellular lysine methyltransferase Set7/9-KMT7 binds HIV-1 TAR RNA, monomethylates the viral transactivator Tat, and enhances HIV transcription.
|
| |
Cell Host Microbe,
7,
234-244.
|
 |
|
|
|
|
 |
T.Umehara,
Y.Nakamura,
M.K.Jang,
K.Nakano,
A.Tanaka,
K.Ozato,
B.Padmanabhan,
and
S.Yokoyama
(2010).
Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain.
|
| |
J Biol Chem,
285,
7610-7618.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Charlop-Powers,
L.Zeng,
Q.Zhang,
and
M.M.Zhou
(2010).
Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2.
|
| |
Cell Res,
20,
529-538.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Huang,
X.D.Yang,
M.M.Zhou,
K.Ozato,
and
L.F.Chen
(2009).
Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA.
|
| |
Mol Cell Biol,
29,
1375-1387.
|
 |
|
|
|
|
 |
F.Vollmuth,
W.Blankenfeldt,
and
M.Geyer
(2009).
Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution.
|
| |
J Biol Chem,
284,
36547-36556.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Thompson
(2009).
Polybromo-1: the chromatin targeting subunit of the PBAF complex.
|
| |
Biochimie,
91,
309-319.
|
 |
|
|
|
|
 |
R.Sanchez,
and
M.M.Zhou
(2009).
The role of human bromodomains in chromatin biology and gene transcription.
|
| |
Curr Opin Drug Discov Devel,
12,
659-665.
|
 |
|
|
|
|
 |
S.Li,
and
M.A.Shogren-Knaak
(2009).
The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes.
|
| |
J Biol Chem,
284,
9411-9417.
|
 |
|
|
|
|
 |
V.W.Gautier,
L.Gu,
N.O'Donoghue,
S.Pennington,
N.Sheehy,
and
W.W.Hall
(2009).
In vitro nuclear interactome of the HIV-1 Tat protein.
|
| |
Retrovirology,
6,
47.
|
 |
|
|
|
|
 |
C.Kupitz,
R.Chandrasekaran,
and
M.Thompson
(2008).
Kinetic analysis of acetylation-dependent Pb1 bromodomain-histone interactions.
|
| |
Biophys Chem,
136,
7.
|
 |
|
|
|
|
 |
H.S.Kwon,
M.M.Brent,
R.Getachew,
P.Jayakumar,
L.F.Chen,
M.Schnolzer,
M.W.McBurney,
R.Marmorstein,
W.C.Greene,
and
M.Ott
(2008).
Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation.
|
| |
Cell Host Microbe,
3,
158-167.
|
 |
|
|
|
|
 |
L.Zeng,
Q.Zhang,
G.Gerona-Navarro,
N.Moshkina,
and
M.M.Zhou
(2008).
Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300.
|
| |
Structure,
16,
643-652.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Thompson,
and
R.Chandrasekaran
(2008).
Thermodynamic analysis of acetylation-dependent Pb1 bromodomain-histone H3 interactions.
|
| |
Anal Biochem,
374,
304-312.
|
 |
|
|
|
|
 |
R.Van Duyne,
K.Kehn-Hall,
Z.Klase,
R.Easley,
M.Heydarian,
M.Saifuddin,
W.Wu,
and
F.Kashanchi
(2008).
Retroviral proteomics and interactomes: intricate balances of cell survival and viral replication.
|
| |
Expert Rev Proteomics,
5,
507-528.
|
 |
|
|
|
|
 |
S.Mujtaba,
K.L.Manzur,
J.R.Gurnon,
M.Kang,
J.L.Van Etten,
and
M.M.Zhou
(2008).
Epigenetic transcriptional repression of cellular genes by a viral SET protein.
|
| |
Nat Cell Biol,
10,
1114-1122.
|
 |
|
|
|
|
 |
T.Hou,
S.Ray,
C.Lee,
and
A.R.Brasier
(2008).
The STAT3 NH2-terminal Domain Stabilizes Enhanceosome Assembly by Interacting with the p300 Bromodomain.
|
| |
J Biol Chem,
283,
30725-30734.
|
 |
|
|
|
|
 |
W.Cao,
C.Bao,
E.Padalko,
and
C.J.Lowenstein
(2008).
Acetylation of mitogen-activated protein kinase phosphatase-1 inhibits Toll-like receptor signaling.
|
| |
J Exp Med,
205,
1491-1503.
|
 |
|
|
|
|
 |
A.Sharma,
S.Awasthi,
C.K.Harrod,
E.F.Matlock,
S.Khan,
L.Xu,
S.Chan,
H.Yang,
C.K.Thammavaram,
R.A.Rasor,
D.K.Burns,
D.J.Skiest,
C.Van Lint,
A.M.Girard,
M.McGee,
R.J.Monnat,
and
R.Harrod
(2007).
The Werner syndrome helicase is a cofactor for HIV-1 long terminal repeat transactivation and retroviral replication.
|
| |
J Biol Chem,
282,
12048-12057.
|
 |
|
|
|
|
 |
C.Hetzer,
D.Bisgrove,
M.S.Cohen,
A.Pedal,
K.Kaehlcke,
A.Speyerer,
K.Bartscherer,
J.Taunton,
and
M.Ott
(2007).
Recruitment and activation of RSK2 by HIV-1 Tat.
|
| |
PLoS ONE,
2,
e151.
|
 |
|
|
|
|
 |
D.M.Heery,
and
P.M.Fischer
(2007).
Pharmacological targeting of lysine acetyltransferases in human disease: a progress report.
|
| |
Drug Discov Today,
12,
88-99.
|
 |
|
|
|
|
 |
H.Huang,
J.Zhang,
W.Shen,
X.Wang,
J.Wu,
J.Wu,
and
Y.Shi
(2007).
Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails.
|
| |
BMC Struct Biol,
7,
57.
|
 |
|
|
|
|
 |
M.Singh,
G.M.Popowicz,
M.Krajewski,
and
T.A.Holak
(2007).
Structural ramification for acetyl-lysine recognition by the bromodomain of human BRG1 protein, a central ATPase of the SWI/SNF remodeling complex.
|
| |
Chembiochem,
8,
1308-1316.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Mujtaba,
L.Zeng,
and
M.M.Zhou
(2007).
Structure and acetyl-lysine recognition of the bromodomain.
|
| |
Oncogene,
26,
5521-5527.
|
 |
|
|
|
|
 |
Y.Nakamura,
T.Umehara,
K.Nakano,
M.K.Jang,
M.Shirouzu,
S.Morita,
H.Uda-Tochio,
H.Hamana,
T.Terada,
N.Adachi,
T.Matsumoto,
A.Tanaka,
M.Horikoshi,
K.Ozato,
B.Padmanabhan,
and
S.Yokoyama
(2007).
Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4.
|
| |
J Biol Chem,
282,
4193-4201.
|
 |
|
|
|
|
 |
A.H.Hassan,
S.Awad,
and
P.Prochasson
(2006).
The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes.
|
| |
J Biol Chem,
281,
18126-18134.
|
 |
|
|
|
|
 |
C.Tréand,
I.du Chéné,
V.Brès,
R.Kiernan,
R.Benarous,
M.Benkirane,
and
S.Emiliani
(2006).
Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter.
|
| |
EMBO J,
25,
1690-1699.
|
 |
|
|
|
|
 |
M.Stevens,
E.De Clercq,
and
J.Balzarini
(2006).
The regulation of HIV-1 transcription: molecular targets for chemotherapeutic intervention.
|
| |
Med Res Rev,
26,
595-625.
|
 |
|
|
|
|
 |
R.Berro,
K.Kehn,
C.de la Fuente,
A.Pumfery,
R.Adair,
J.Wade,
A.M.Colberg-Poley,
J.Hiscott,
and
F.Kashanchi
(2006).
Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32.
|
| |
J Virol,
80,
3189-3204.
|
 |
|
|
|
|
 |
S.Pantano,
A.Marcello,
A.Ferrari,
D.Gaudiosi,
A.Sabò,
V.Pellegrini,
F.Beltram,
M.Giacca,
and
P.Carloni
(2006).
Insights on HIV-1 Tat:P/CAF bromodomain molecular recognition from in vivo experiments and molecular dynamics simulations.
|
| |
Proteins,
62,
1062-1073.
|
 |
|
|
|
|
 |
S.Shojania,
and
J.D.O'Neil
(2006).
HIV-1 Tat is a natively unfolded protein: the solution conformation and dynamics of reduced HIV-1 Tat-(1-72) by NMR spectroscopy.
|
| |
J Biol Chem,
281,
8347-8356.
|
 |
|
|
|
|
 |
T.Mahmoudi,
M.Parra,
R.G.Vries,
S.E.Kauder,
C.P.Verrijzer,
M.Ott,
and
E.Verdin
(2006).
The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter.
|
| |
J Biol Chem,
281,
19960-19968.
|
 |
|
|
|
|
 |
K.Wong,
A.Sharma,
S.Awasthi,
E.F.Matlock,
L.Rogers,
C.Van Lint,
D.J.Skiest,
D.K.Burns,
and
R.Harrod
(2005).
HIV-1 Tat interactions with p300 and PCAF transcriptional coactivators inhibit histone acetylation and neurotrophin signaling through CREB.
|
| |
J Biol Chem,
280,
9390-9399.
|
 |
|
|
|
|
 |
L.Li,
H.S.Li,
C.D.Pauza,
M.Bukrinsky,
and
R.Y.Zhao
(2005).
Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions.
|
| |
Cell Res,
15,
923-934.
|
 |
|
|
|
|
 |
S.Amini,
G.Mameli,
L.Del Valle,
A.Skowronska,
K.Reiss,
B.B.Gelman,
M.K.White,
K.Khalili,
and
B.E.Sawaya
(2005).
p73 Interacts with human immunodeficiency virus type 1 Tat in astrocytic cells and prevents its acetylation on lysine 28.
|
| |
Mol Cell Biol,
25,
8126-8138.
|
 |
|
|
|
|
 |
S.Pagans,
A.Pedal,
B.J.North,
K.Kaehlcke,
B.L.Marshall,
A.Dorr,
C.Hetzer-Egger,
P.Henklein,
R.Frye,
M.W.McBurney,
H.Hruby,
M.Jung,
E.Verdin,
and
M.Ott
(2005).
SIRT1 regulates HIV transcription via Tat deacetylation.
|
| |
PLoS Biol,
3,
e41.
|
 |
|
|
|
|
 |
S.Pantano,
and
P.Carloni
(2005).
Comparative analysis of HIV-1 Tat variants.
|
| |
Proteins,
58,
638-643.
|
 |
|
|
|
|
 |
Y.Desfosses,
M.Solis,
Q.Sun,
N.Grandvaux,
C.Van Lint,
A.Burny,
A.Gatignol,
M.A.Wainberg,
R.Lin,
and
J.Hiscott
(2005).
Regulation of human immunodeficiency virus type 1 gene expression by clade-specific Tat proteins.
|
| |
J Virol,
79,
9180-9191.
|
 |
|
|
|
|
 |
M.J.Bottomley
(2004).
Structures of protein domains that create or recognize histone modifications.
|
| |
EMBO Rep,
5,
464-469.
|
 |
|
|
|
|
 |
M.Kasten,
H.Szerlong,
H.Erdjument-Bromage,
P.Tempst,
M.Werner,
and
B.R.Cairns
(2004).
Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14.
|
| |
EMBO J,
23,
1348-1359.
|
 |
|
|
|
|
 |
S.Mujtaba,
Y.He,
L.Zeng,
S.Yan,
O.Plotnikova,
Sachchidanand,
R.Sanchez,
N.J.Zeleznik-Le,
Z.Ronai,
and
M.M.Zhou
(2004).
Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
|
| |
Mol Cell,
13,
251-263.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.J.Yang
(2004).
The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases.
|
| |
Nucleic Acids Res,
32,
959-976.
|
 |
|
|
|
|
 |
X.J.Yang
(2004).
Lysine acetylation and the bromodomain: a new partnership for signaling.
|
| |
Bioessays,
26,
1076-1087.
|
 |
|
|
|
|
 |
C.Brigati,
M.Giacca,
D.M.Noonan,
and
A.Albini
(2003).
HIV Tat, its TARgets and the control of viral gene expression.
|
| |
FEMS Microbiol Lett,
220,
57-65.
|
 |
|
|
|
|
 |
C.Caron,
E.Col,
and
S.Khochbin
(2003).
The viral control of cellular acetylation signaling.
|
| |
Bioessays,
25,
58-65.
|
 |
|
|
|
|
 |
C.Pivot-Pajot,
C.Caron,
J.Govin,
A.Vion,
S.Rousseaux,
and
S.Khochbin
(2003).
Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein.
|
| |
Mol Cell Biol,
23,
5354-5365.
|
 |
|
|
|
|
 |
K.Kaehlcke,
A.Dorr,
C.Hetzer-Egger,
V.Kiermer,
P.Henklein,
M.Schnoelzer,
E.Loret,
P.A.Cole,
E.Verdin,
and
M.Ott
(2003).
Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation.
|
| |
Mol Cell,
12,
167-176.
|
 |
|
|
|
|
 |
K.Strebel
(2003).
Virus-host interactions: role of HIV proteins Vif, Tat, and Rev.
|
| |
AIDS,
17,
S25-S34.
|
 |
|
|
|
|
 |
M.Lusic,
A.Marcello,
A.Cereseto,
and
M.Giacca
(2003).
Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter.
|
| |
EMBO J,
22,
6550-6561.
|
 |
|
|
|
|
 |
R.Harrod,
J.Nacsa,
C.Van Lint,
J.Hansen,
T.Karpova,
J.McNally,
and
G.Franchini
(2003).
Human immunodeficiency virus type-1 Tat/co-activator acetyltransferase interactions inhibit p53Lys-320 acetylation and p53-responsive transcription.
|
| |
J Biol Chem,
278,
12310-12318.
|
 |
|
|
|
|
 |
T.Pawson,
and
P.Nash
(2003).
Assembly of cell regulatory systems through protein interaction domains.
|
| |
Science,
300,
445-452.
|
 |
|
|
|
|
 |
W.K.Wang,
V.Tereshko,
P.Boccuni,
D.MacGrogan,
S.D.Nimer,
and
D.J.Patel
(2003).
Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
|
| |
Structure,
11,
775-789.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Dorr,
V.Kiermer,
A.Pedal,
H.R.Rackwitz,
P.Henklein,
U.Schubert,
M.M.Zhou,
E.Verdin,
and
M.Ott
(2002).
Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain.
|
| |
EMBO J,
21,
2715-2723.
|
 |
|
|
|
|
 |
E.Col,
B.Gilquin,
C.Caron,
and
S.Khochbin
(2002).
Tat-controlled protein acetylation.
|
| |
J Biol Chem,
277,
37955-37960.
|
 |
|
|
|
|
 |
E.Remboutsika,
K.Yamamoto,
M.Harbers,
and
M.Schmutz
(2002).
The bromodomain mediates transcriptional intermediary factor 1alpha -nucleosome interactions.
|
| |
J Biol Chem,
277,
50318-50325.
|
 |
|
|
|
|
 |
R.G.Ptak
(2002).
HIV-1 regulatory proteins: targets for novel drug development.
|
| |
Expert Opin Investig Drugs,
11,
1099-1115.
|
 |
|
|
|
|
 |
V.Brès,
H.Tagami,
J.M.Péloponèse,
E.Loret,
K.T.Jeang,
Y.Nakatani,
S.Emiliani,
M.Benkirane,
and
R.E.Kiernan
(2002).
Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF.
|
| |
EMBO J,
21,
6811-6819.
|
 |
|
|
|
|
 |
Y.Nakatani
(2002).
HIV-1 transcription: activation mediated by acetylation of Tat.
|
| |
Structure,
10,
443-444.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

