 |
PDBsum entry 1jba

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.6.1.2
- guanylate cyclase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
GTP = 3',5'-cyclic GMP + diphosphate
|
 |
 |
 |
 |
 |
 GTP
GTP
|
=
|
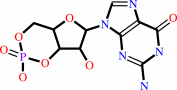 3',5'-cyclic GMP
3',5'-cyclic GMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
274:19329-19337
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases.
|
|
J.B.Ames,
A.M.Dizhoor,
M.Ikura,
K.Palczewski,
L.Stryer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Guanylyl cyclase activating protein-2 (GCAP-2) is a Ca2+-sensitive regulator of
phototransduction in retinal photoreceptor cells. GCAP-2 activates retinal
guanylyl cyclases at low Ca2+ concentration (<100 nM) and inhibits them at
high Ca2+ (>500 nM). The light-induced lowering of the Ca2+ level from
approximately 500 nM in the dark to approximately 50 nM following illumination
is known to play a key role in visual recovery and adaptation. We report here
the three-dimensional structure of unmyristoylated GCAP-2 with three bound Ca2+
ions as determined by nuclear magnetic resonance spectroscopy of recombinant,
isotopically labeled protein. GCAP-2 contains four EF-hand motifs arranged in a
compact tandem array like that seen previously in recoverin. The root mean
square deviation of the main chain atoms in the EF-hand regions is 2.2 A in
comparing the Ca2+-bound structures of GCAP-2 and recoverin. EF-1, as in
recoverin, does not bind calcium because it contains a disabling Cys-Pro
sequence. GCAP-2 differs from recoverin in that the calcium ion binds to EF-4 in
addition to EF-2 and EF-3. A prominent exposed patch of hydrophobic residues
formed by EF-1 and EF-2 (Leu24, Trp27, Phe31, Phe45, Phe48, Phe49, Tyr81, Val82,
Leu85, and Leu89) may serve as a target-binding site for the transmission of
calcium signals to guanylyl cyclase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Fig. 5. Schematic ribbon representation (A) and
space-filling model (B) of the energy-minimized average
structure of unmyristoylated GCAP-2 with three Ca^2+ bound. The
side chain atoms of residues at the domain interface (Ala^63,
Ala^67, Ile^103, and Ile^120) are shown in A and the color
scheme is as in Fig. 4. The figure was generated using Molscript
(49) and Raster3d (23).
|
 |
Figure 8.
Fig. 8. Space-filling representation (A) and
ball-and-stick model (B) of side chain atoms of the exposed
hydrophobic patch of GCAP-2. Hydrophobic, negatively charged,
and positively charged residues are highlighted in yellow, red,
and blue, respectively. Solvent-exposed hydrophobic residues
from EF-1 and EF-2 are indicated.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(1999,
274,
19329-19337)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Theisgen,
L.Thomas,
T.Schröder,
C.Lange,
M.Kovermann,
J.Balbach,
and
D.Huster
(2011).
The presence of membranes or micelles induces structural changes of the myristoylated guanylate-cyclase activating protein-2.
|
| |
Eur Biophys J,
40,
565-576.
|
 |
|
|
|
|
 |
A.M.Dizhoor,
E.V.Olshevskaya,
and
I.V.Peshenko
(2010).
Mg2+/Ca2+ cation binding cycle of guanylyl cyclase activating proteins (GCAPs): role in regulation of photoreceptor guanylyl cyclase.
|
| |
Mol Cell Biochem,
334,
117-124.
|
 |
|
|
|
|
 |
P.Behnen,
A.Scholten,
N.Rätscho,
and
K.W.Koch
(2009).
The cone-specific calcium sensor guanylate cyclase activating protein 4 from the zebrafish retina.
|
| |
J Biol Inorg Chem,
14,
89-99.
|
 |
|
|
|
|
 |
S.Lim,
I.Peshenko,
A.Dizhoor,
and
J.B.Ames
(2009).
Effects of Ca2+, Mg2+, and myristoylation on guanylyl cyclase activating protein 1 structure and stability.
|
| |
Biochemistry,
48,
850-862.
|
 |
|
|
|
|
 |
I.V.Peshenko,
E.V.Olshevskaya,
and
A.M.Dizhoor
(2008).
Binding of guanylyl cyclase activating protein 1 (GCAP1) to retinal guanylyl cyclase (RetGC1). The role of individual EF-hands.
|
| |
J Biol Chem,
283,
21747-21757.
|
 |
|
|
|
|
 |
R.Stephen,
S.Filipek,
K.Palczewski,
and
M.C.Sousa
(2008).
Ca2+ -dependent regulation of phototransduction.
|
| |
Photochem Photobiol,
84,
903-910.
|
 |
|
|
|
|
 |
I.V.Peshenko,
and
A.M.Dizhoor
(2007).
Activation and inhibition of photoreceptor guanylyl cyclase by guanylyl cyclase activating protein 1 (GCAP-1): the functional role of Mg2+/Ca2+ exchange in EF-hand domains.
|
| |
J Biol Chem,
282,
21645-21652.
|
 |
|
|
|
|
 |
R.D.Burgoyne
(2007).
Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling.
|
| |
Nat Rev Neurosci,
8,
182-193.
|
 |
|
|
|
|
 |
R.Stephen,
G.Bereta,
M.Golczak,
K.Palczewski,
and
M.C.Sousa
(2007).
Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1.
|
| |
Structure,
15,
1392-1402.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Sokal,
W.J.Dupps,
M.A.Grassi,
J.Brown,
L.M.Affatigato,
N.Roychowdhury,
L.Yang,
S.Filipek,
K.Palczewski,
E.M.Stone,
and
W.Baehr
(2005).
A novel GCAP1 missense mutation (L151F) in a large family with autosomal dominant cone-rod dystrophy (adCORD).
|
| |
Invest Ophthalmol Vis Sci,
46,
1124-1132.
|
 |
|
|
|
|
 |
J.N.Wingard,
J.Chan,
I.Bosanac,
F.Haeseleer,
K.Palczewski,
M.Ikura,
and
J.B.Ames
(2005).
Structural analysis of Mg2+ and Ca2+ binding to CaBP1, a neuron-specific regulator of calcium channels.
|
| |
J Biol Chem,
280,
37461-37470.
|
 |
|
|
|
|
 |
P.I.Lario,
R.A.Pfuetzner,
E.A.Frey,
L.Creagh,
C.Haynes,
A.T.Maurelli,
and
N.C.Strynadka
(2005).
Structure and biochemical analysis of a secretin pilot protein.
|
| |
EMBO J,
24,
1111-1121.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Y.Hwang,
R.Schlesinger,
and
K.W.Koch
(2004).
Irregular dimerization of guanylate cyclase-activating protein 1 mutants causes loss of target activation.
|
| |
Eur J Biochem,
271,
3785-3793.
|
 |
|
|
|
|
 |
K.M.Nishiguchi,
I.Sokal,
L.Yang,
N.Roychowdhury,
K.Palczewski,
E.L.Berson,
T.P.Dryja,
and
W.Baehr
(2004).
A novel mutation (I143NT) in guanylate cyclase-activating protein 1 (GCAP1) associated with autosomal dominant cone degeneration.
|
| |
Invest Ophthalmol Vis Sci,
45,
3863-3870.
|
 |
|
|
|
|
 |
W.Zhou,
Y.Qian,
K.Kunjilwar,
P.J.Pfaffinger,
and
S.Choe
(2004).
Structural insights into the functional interaction of KChIP1 with Shal-type K(+) channels.
|
| |
Neuron,
41,
573-586.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Imanishi,
L.Yang,
I.Sokal,
S.Filipek,
K.Palczewski,
and
W.Baehr
(2004).
Diversity of guanylate cyclase-activating proteins (GCAPs) in teleost fish: characterization of three novel GCAPs (GCAP4, GCAP5, GCAP7) from zebrafish (Danio rerio) and prediction of eight GCAPs (GCAP1-8) in pufferfish (Fugu rubripes).
|
| |
J Mol Evol,
59,
204-217.
|
 |
|
|
|
|
 |
G.M.Pitari,
L.V.Zingman,
D.M.Hodgson,
A.E.Alekseev,
S.Kazerounian,
M.Bienengraeber,
G.Hajnóczky,
A.Terzic,
and
S.A.Waldman
(2003).
Bacterial enterotoxins are associated with resistance to colon cancer.
|
| |
Proc Natl Acad Sci U S A,
100,
2695-2699.
|
 |
|
|
|
|
 |
I.Sokal,
A.Alekseev,
and
K.Palczewski
(2003).
Photoreceptor guanylate cyclase variants: cGMP production under control.
|
| |
Acta Biochim Pol,
50,
1075-1095.
|
 |
|
|
|
|
 |
K.D.Ridge,
N.G.Abdulaev,
M.Sousa,
and
K.Palczewski
(2003).
Phototransduction: crystal clear.
|
| |
Trends Biochem Sci,
28,
479-487.
|
 |
|
|
|
|
 |
T.P.Sakmar,
S.T.Menon,
E.P.Marin,
and
E.S.Awad
(2002).
Rhodopsin: insights from recent structural studies.
|
| |
Annu Rev Biophys Biomol Struct,
31,
443-484.
|
 |
|
|
|
|
 |
Y.Imanishi,
N.Li,
I.Sokal,
M.E.Sowa,
O.Lichtarge,
T.G.Wensel,
D.A.Saperstein,
W.Baehr,
and
K.Palczewski
(2002).
Characterization of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man.
|
| |
Eur J Neurosci,
15,
63-78.
|
 |
|
|
|
|
 |
M.Palczewska,
P.Groves,
A.Ambrus,
A.Kaleta,
K.E.Kövér,
G.Batta,
and
J.Kuźnicki
(2001).
Structural and biochemical characterization of neuronal calretinin domain I-II (residues 1-100). Comparison to homologous calbindin D28k domain I-II (residues 1-93).
|
| |
Eur J Biochem,
268,
6229-6237.
|
 |
|
|
|
|
 |
N.Li,
I.Sokal,
J.D.Bronson,
K.Palczewski,
and
W.Baehr
(2001).
Identification of functional regions of guanylate cyclase-activating protein 1 (GCAP1) using GCAP1/GCIP chimeras.
|
| |
Biol Chem,
382,
1179-1188.
|
 |
|
|
|
|
 |
A.Lewit-Bentley,
and
S.Réty
(2000).
EF-hand calcium-binding proteins.
|
| |
Curr Opin Struct Biol,
10,
637-643.
|
 |
|
|
|
|
 |
F.Haeseleer,
I.Sokal,
C.L.Verlinde,
H.Erdjument-Bromage,
P.Tempst,
A.N.Pronin,
J.L.Benovic,
R.N.Fariss,
and
K.Palczewski
(2000).
Five members of a novel Ca(2+)-binding protein (CABP) subfamily with similarity to calmodulin.
|
| |
J Biol Chem,
275,
1247-1260.
|
 |
|
|
|
|
 |
J.B.Ames,
K.B.Hendricks,
T.Strahl,
I.G.Huttner,
N.Hamasaki,
and
J.Thorner
(2000).
Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae.
|
| |
Biochemistry,
39,
12149-12161.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Palczewski,
A.S.Polans,
W.Baehr,
and
J.B.Ames
(2000).
Ca(2+)-binding proteins in the retina: structure, function, and the etiology of human visual diseases.
|
| |
Bioessays,
22,
337-350.
|
 |
|
|
|
|
 |
R.D.Hamer
(2000).
Computational analysis of vertebrate phototransduction: combined quantitative and qualitative modeling of dark- and light-adapted responses in amphibian rods.
|
| |
Vis Neurosci,
17,
679-699.
|
 |
|
|
|
|
 |
K.L.Yap,
J.B.Ames,
M.B.Swindells,
and
M.Ikura
(1999).
Diversity of conformational states and changes within the EF-hand protein superfamily.
|
| |
Proteins,
37,
499-507.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

