 |
PDBsum entry 1e8h

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1e8h
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.3.38
- vanillyl-alcohol oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxy-3-methoxy-benzenemethanol + O2 = vanillin + H2O2
|
 |
 |
 |
 |
 |
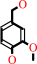 4-hydroxy-3-methoxy-benzenemethanol
4-hydroxy-3-methoxy-benzenemethanol
|
+
|
 O2
O2
|
=
|
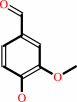 vanillin
vanillin
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
ADP)
matches with 50.94% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
275:38654-38658
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural analysis of flavinylation in vanillyl-alcohol oxidase.
|
|
M.W.Fraaije,
R.H.van Den Heuvel,
W.J.van Berkel,
A.Mattevi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Vanillyl-alcohol oxidase (VAO) is member of a newly recognized flavoprotein
family of structurally related oxidoreductases. The enzyme contains a covalently
linked FAD cofactor. To study the mechanism of flavinylation we have created a
design point mutation (His-61 --> Thr). In the mutant enzyme the covalent
His-C8alpha-flavin linkage is not formed, while the enzyme is still able to bind
FAD and perform catalysis. The H61T mutant displays a similar affinity for FAD
and ADP (K(d) = 1.8 and 2.1 microm, respectively) but does not interact with
FMN. H61T is about 10-fold less active with 4-(methoxymethyl)phenol) (k(cat) =
0.24 s(-)(1), K(m) = 40 microm) than the wild-type enzyme. The crystal
structures of both the holo and apo form of H61T are highly similar to the
structure of wild-type VAO, indicating that binding of FAD to the apoprotein
does not require major structural rearrangements. These results show that
covalent flavinylation is an autocatalytical process in which His-61 plays a
crucial role by activating His-422. Furthermore, our studies clearly demonstrate
that in VAO, the FAD binds via a typical lock-and-key approach to a preorganized
binding site.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. The active site of the H61T ternary complex
structure showing all relevant residues, the isoalloxazine ring
of the FAD cofactor (yellow) and the bound
4-(trifluoromethyl)phenol (blue) contoured by the 2F[o]  F[c]
electron density calculated with the refined model. The picture
was generated with DINO (27). F[c]
electron density calculated with the refined model. The picture
was generated with DINO (27).
|
 |
Figure 3.
Fig. 3. A, a section through the C  trace of
the apoenzyme subunit showing the large internal solvent-filled
cavity. For clarity, the FAD cofactor and
4-(trifluoromethyl)phenol as bound in the holo H61T ternary
complex were superimposed and shown in yellow and blue,
respectively. The surface was calculated using MSMS (28) and the
picture was generated using DINO (27). B, superposition of
active site residues in the apo (green) and the holo form
complexed with 4-(trifluoromethyl)phenol (red) structures of the
VAO H61T mutant. The figure was prepared with MOLSCRIPT ( 26). trace of
the apoenzyme subunit showing the large internal solvent-filled
cavity. For clarity, the FAD cofactor and
4-(trifluoromethyl)phenol as bound in the holo H61T ternary
complex were superimposed and shown in yellow and blue,
respectively. The surface was calculated using MSMS (28) and the
picture was generated using DINO (27). B, superposition of
active site residues in the apo (green) and the holo form
complexed with 4-(trifluoromethyl)phenol (red) structures of the
VAO H61T mutant. The figure was prepared with MOLSCRIPT ( 26).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2000,
275,
38654-38658)
copyright 2000.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.Fincato,
P.N.Moschou,
V.Spedaletti,
R.Tavazza,
R.Angelini,
R.Federico,
K.A.Roubelakis-Angelakis,
and
P.Tavladoraki
(2011).
Functional diversity inside the Arabidopsis polyamine oxidase gene family.
|
| |
J Exp Bot,
62,
1155-1168.
|
 |
|
|
|
|
 |
D.P.Heuts,
N.S.Scrutton,
W.S.McIntire,
and
M.W.Fraaije
(2009).
What's in a covalent bond? On the role and formation of covalently bound flavin cofactors.
|
| |
FEBS J,
276,
3405-3427.
|
 |
|
|
|
|
 |
C.H.Huang,
A.Winkler,
C.L.Chen,
W.L.Lai,
Y.C.Tsai,
P.Macheroux,
and
S.H.Liaw
(2008).
Functional roles of the 6-S-cysteinyl, 8alpha-N1-histidyl FAD in glucooligosaccharide oxidase from Acremonium strictum.
|
| |
J Biol Chem,
283,
30990-30996.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Jin,
H.Mazon,
R.H.van den Heuvel,
A.J.Heck,
D.B.Janssen,
and
M.W.Fraaije
(2008).
Covalent flavinylation of vanillyl-alcohol oxidase is an autocatalytic process.
|
| |
FEBS J,
275,
5191-5200.
|
 |
|
|
|
|
 |
N.G.Leferink,
W.A.van den Berg,
and
W.J.van Berkel
(2008).
l-Galactono-gamma-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis.
|
| |
FEBS J,
275,
713-726.
|
 |
|
|
|
|
 |
I.Alexeev,
A.Sultana,
P.Mäntsälä,
J.Niemi,
and
G.Schneider
(2007).
Aclacinomycin oxidoreductase (AknOx) from the biosynthetic pathway of the antibiotic aclacinomycin is an unusual flavoenzyme with a dual active site.
|
| |
Proc Natl Acad Sci U S A,
104,
6170-6175.
|
 |
|
|
|
|
 |
J.Jin,
H.Mazon,
R.H.van den Heuvel,
D.B.Janssen,
and
M.W.Fraaije
(2007).
Discovery of a eugenol oxidase from Rhodococcus sp. strain RHA1.
|
| |
FEBS J,
274,
2311-2321.
|
 |
|
|
|
|
 |
N.Masuoka,
K.Nihei,
and
I.Kubo
(2006).
Xanthine oxidase inhibitory activity of alkyl gallates.
|
| |
Mol Nutr Food Res,
50,
725-731.
|
 |
|
|
|
|
 |
A.Hassan-Abdallah,
R.C.Bruckner,
G.Zhao,
and
M.S.Jorns
(2005).
Biosynthesis of covalently bound flavin: isolation and in vitro flavinylation of the monomeric sarcosine oxidase apoprotein.
|
| |
Biochemistry,
44,
6452-6462.
|
 |
|
|
|
|
 |
S.Anderson,
V.Dragnea,
S.Masuda,
J.Ybe,
K.Moffat,
and
C.Bauer
(2005).
Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides.
|
| |
Biochemistry,
44,
7998-8005.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.J.Heck,
and
R.H.Van Den Heuvel
(2004).
Investigation of intact protein complexes by mass spectrometry.
|
| |
Mass Spectrom Rev,
23,
368-389.
|
 |
|
|
|
|
 |
C.B.Chiribau,
C.Sandu,
M.Fraaije,
E.Schiltz,
and
R.Brandsch
(2004).
A novel gamma-N-methylaminobutyrate demethylating oxidase involved in catabolism of the tobacco alkaloid nicotine by Arthrobacter nicotinovorans pAO1.
|
| |
Eur J Biochem,
271,
4677-4684.
|
 |
|
|
|
|
 |
G.Cecchini
(2003).
Function and structure of complex II of the respiratory chain.
|
| |
Annu Rev Biochem,
72,
77.
|
 |
|
|
|
|
 |
M.H.Hefti,
J.Vervoort,
and
W.J.van Berkel
(2003).
Deflavination and reconstitution of flavoproteins.
|
| |
Eur J Biochem,
270,
4227-4242.
|
 |
|
|
|
|
 |
R.T.Bossi,
A.Negri,
G.Tedeschi,
and
A.Mattevi
(2002).
Structure of FAD-bound L-aspartate oxidase: insight into substrate specificity and catalysis.
|
| |
Biochemistry,
41,
3018-3024.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Eschenbrenner,
L.J.Chlumsky,
P.Khanna,
F.Strasser,
and
M.S.Jorns
(2001).
Organization of the multiple coenzymes and subunits and role of the covalent flavin link in the complex heterotetrameric sarcosine oxidase.
|
| |
Biochemistry,
40,
5352-5367.
|
 |
|
|
|
|
 |
N.Tahallah,
M.Pinkse,
C.S.Maier,
and
A.J.Heck
(2001).
The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument.
|
| |
Rapid Commun Mass Spectrom,
15,
596-601.
|
 |
|
|
|
|
 |
O.Dym,
and
D.Eisenberg
(2001).
Sequence-structure analysis of FAD-containing proteins.
|
| |
Protein Sci,
10,
1712-1728.
|
 |
|
|
|
|
 |
S.Crosson,
and
K.Moffat
(2001).
Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction.
|
| |
Proc Natl Acad Sci U S A,
98,
2995-3000.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

