 |
PDBsum entry 1bk0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
B-lactam antibiotic
|
PDB id
|
|

|

|
1bk0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.21.3.1
- isopenicillin-N synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Penicillin N and Deacetoxycephalosporin C Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine + O2 = isopenicillin N + 2 H2O
|
 |
 |
 |
 |
 |
N-[(5S)-5-amino-5-carboxypentanoyl]-L-cysteinyl-D-valine
|
+
|
 O2
O2
|
=
|
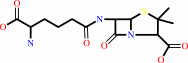 isopenicillin N
isopenicillin N
|
+
|
2
×
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
387:827-830
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation.
|
|
P.L.Roach,
I.J.Clifton,
C.M.Hensgens,
N.Shibata,
C.J.Schofield,
J.Hajdu,
J.E.Baldwin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The biosynthesis of penicillin and cephalosporin antibiotics in microorganisms
requires the formation of the bicyclic nucleus of penicillin. Isopenicillin N
synthase (IPNS), a non-haem iron-dependent oxidase, catalyses the reaction of a
tripeptide, delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-valine (ACV), and
dioxygen to form isopenicillin N and two water molecules. Mechanistic studies
suggest the reaction is initiated by ligation of the substrate thiolate to the
iron centre, and proceeds through an enzyme-bound monocyclic intermediate. Here
we report the crystal structure of IPNS complexed to ferrous iron and ACV,
determined to 1.3 A resolution. Based on the structure, we propose a mechanism
for penicillin formation that involves ligation of ACV to the iron centre,
creating a vacant iron coordination site into which dioxygen can bind.
Subsequently, iron-dioxygen and iron-oxo species remove the requisite hydrogens
from ACV without the direct assistance of protein residues. The crystal
structure of the complex with the dioxygen analogue, NO and ACV bound to the
active-site iron supports this hypothesis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2 Mechanism for isopenicillin N formation and the
formation of the Fe: ACV: NO:. sp;IPNS complex.
|
 |
Figure 3.
Figure 3 Comparison of the structures of Mn: IPNS (a) and
Fe(II): ACV: IPNS (. b) from the same orientation. The
jelly-roll motif is in green, the C-terminal region (residues
313-331) cyan, the active-site metal ion (manganese in a, iron
in b) orange, the key substrate-binding residues (His 214, His
270, Asp 216, Arg 87, Arg 279, Tyr 189 and Ser 281) magenta, and
the ACV yellow.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nature
(1997,
387,
827-830)
copyright 1997.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.K.Sydor,
S.M.Barry,
O.M.Odulate,
F.Barona-Gomez,
S.W.Haynes,
C.Corre,
L.Song,
and
G.L.Challis
(2011).
Regio- and stereodivergent antibiotic oxidative carbocyclizations catalysed by Rieske oxygenase-like enzymes.
|
| |
Nat Chem,
3,
388-392.
|
 |
|
|
|
|
 |
W.A.Schenk
(2011).
The coordination chemistry of small sulfur-containing molecules: a personal perspective.
|
| |
Dalton Trans,
40,
1209-1219.
|
 |
|
|
|
|
 |
C.D.Brown-Marshall,
A.R.Diebold,
and
E.I.Solomon
(2010).
Reaction coordinate of isopenicillin N synthase: oxidase versus oxygenase activity.
|
| |
Biochemistry,
49,
1176-1182.
|
 |
|
|
|
|
 |
C.T.Walsh,
and
M.A.Fischbach
(2010).
Natural products version 2.0: connecting genes to molecules.
|
| |
J Am Chem Soc,
132,
2469-2493.
|
 |
|
|
|
|
 |
F.Dahmani-Mardas,
C.Troadec,
A.Boualem,
S.Lévêque,
A.A.Alsadon,
A.A.Aldoss,
C.Dogimont,
and
A.Bendahmane
(2010).
Engineering melon plants with improved fruit shelf life using the TILLING approach.
|
| |
PLoS One,
5,
e15776.
|
 |
|
|
|
|
 |
L.M.Blank,
B.E.Ebert,
K.Buehler,
and
B.Bühler
(2010).
Redox biocatalysis and metabolism: molecular mechanisms and metabolic network analysis.
|
| |
Antioxid Redox Signal,
13,
349-394.
|
 |
|
|
|
|
 |
E.M.Nolan,
and
C.T.Walsh
(2009).
How nature morphs peptide scaffolds into antibiotics.
|
| |
Chembiochem,
10,
34-53.
|
 |
|
|
|
|
 |
K.Morokuma
(2009).
Theoretical studies of structure, function and reactivity of molecules-A personal account.
|
| |
Proc Jpn Acad Ser B Phys Biol Sci,
85,
167-182.
|
 |
|
|
|
|
 |
R.Chowdhury,
M.A.McDonough,
J.Mecinović,
C.Loenarz,
E.Flashman,
K.S.Hewitson,
C.Domene,
and
C.J.Schofield
(2009).
Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases.
|
| |
Structure,
17,
981-989.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Ge,
I.J.Clifton,
A.R.Howard-Jones,
J.E.Stok,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2009).
Structural studies on the reaction of isopenicillin N synthase with a sterically demanding depsipeptide substrate analogue.
|
| |
Chembiochem,
10,
2025-2031.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Andreini,
I.Bertini,
G.Cavallaro,
G.L.Holliday,
and
J.M.Thornton
(2008).
Metal ions in biological catalysis: from enzyme databases to general principles.
|
| |
J Biol Inorg Chem,
13,
1205-1218.
|
 |
|
|
|
|
 |
E.G.Kovaleva,
and
J.D.Lipscomb
(2008).
Versatility of biological non-heme Fe(II) centers in oxygen activation reactions.
|
| |
Nat Chem Biol,
4,
186-193.
|
 |
|
|
|
|
 |
L.M.Mirica,
and
J.P.Klinman
(2008).
The nature of O2 activation by the ethylene-forming enzyme 1-aminocyclopropane-1-carboxylic acid oxidase.
|
| |
Proc Natl Acad Sci U S A,
105,
1814-1819.
|
 |
|
|
|
|
 |
P.C.Bruijnincx,
G.van Koten,
and
R.J.Klein Gebbink
(2008).
Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: recent developments in enzymology and modeling studies.
|
| |
Chem Soc Rev,
37,
2716-2744.
|
 |
|
|
|
|
 |
A.C.Stewart,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2007).
A cyclobutanone analogue mimics penicillin in binding to isopenicillin N synthase.
|
| |
Chembiochem,
8,
2003-2007.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Siddiq,
L.R.Aminova,
and
R.R.Ratan
(2007).
Hypoxia inducible factor prolyl 4-hydroxylase enzymes: center stage in the battle against hypoxia, metabolic compromise and oxidative stress.
|
| |
Neurochem Res,
32,
931-946.
|
 |
|
|
|
|
 |
C.A.Joseph,
and
M.J.Maroney
(2007).
Cysteine dioxygenase: structure and mechanism.
|
| |
Chem Commun (Camb),
(),
3338-3349.
|
 |
|
|
|
|
 |
C.D.Brown,
M.L.Neidig,
M.B.Neibergall,
J.D.Lipscomb,
and
E.I.Solomon
(2007).
VTVH-MCD and DFT studies of thiolate bonding to [FeNO]7/[FeO2]8 complexes of isopenicillin N synthase: substrate determination of oxidase versus oxygenase activity in nonheme Fe enzymes.
|
| |
J Am Chem Soc,
129,
7427-7438.
|
 |
|
|
|
|
 |
E.Flashman,
and
C.J.Schofield
(2007).
The most versatile of all reactive intermediates?
|
| |
Nat Chem Biol,
3,
86-87.
|
 |
|
|
|
|
 |
J.M.Bollinger,
and
C.Krebs
(2007).
Enzymatic C-H activation by metal-superoxo intermediates.
|
| |
Curr Opin Chem Biol,
11,
151-158.
|
 |
|
|
|
|
 |
A.Daruzzaman,
I.J.Clifton,
R.M.Adlington,
J.E.Baldwin,
and
P.J.Rutledge
(2006).
Unexpected oxidation of a depsipeptide substrate analogue in crystalline isopenicillin N synthase.
|
| |
Chembiochem,
7,
351-358.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Schöneich,
and
V.S.Sharov
(2006).
Mass spectrometry of protein modifications by reactive oxygen and nitrogen species.
|
| |
Free Radic Biol Med,
41,
1507-1520.
|
 |
|
|
|
|
 |
L.M.Hoffart,
E.W.Barr,
R.B.Guyer,
J.M.Bollinger,
and
C.Krebs
(2006).
Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase.
|
| |
Proc Natl Acad Sci U S A,
103,
14738-14743.
|
 |
|
|
|
|
 |
A.Karlsson,
J.V.Parales,
R.E.Parales,
D.T.Gibson,
H.Eklund,
and
S.Ramaswamy
(2005).
NO binding to naphthalene dioxygenase.
|
| |
J Biol Inorg Chem,
10,
483-489.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Bitto,
C.A.Bingman,
S.T.Allard,
G.E.Wesenberg,
D.J.Aceti,
R.L.Wrobel,
R.O.Frederick,
H.Sreenath,
F.C.Vojtik,
W.B.Jeon,
C.S.Newman,
J.Primm,
M.R.Sussman,
B.G.Fox,
J.L.Markley,
and
G.N.Phillips
(2005).
The structure at 2.4 A resolution of the protein from gene locus At3g21360, a putative Fe(II)/2-oxoglutarate-dependent enzyme from Arabidopsis thaliana.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
469-472.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.D.Koehntop,
J.P.Emerson,
and
L.Que
(2005).
The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes.
|
| |
J Biol Inorg Chem,
10,
87-93.
|
 |
|
|
|
|
 |
L.J.Higgins,
F.Yan,
P.Liu,
H.W.Liu,
and
C.L.Drennan
(2005).
Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme.
|
| |
Nature,
437,
838-844.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.J.Spiering,
C.D.Moon,
H.H.Wilkinson,
and
C.L.Schardl
(2005).
Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum.
|
| |
Genetics,
169,
1403-1414.
|
 |
|
|
|
|
 |
M.L.Neidig,
and
E.I.Solomon
(2005).
Structure-function correlations in oxygen activating non-heme iron enzymes.
|
| |
Chem Commun (Camb),
(),
5843-5863.
|
 |
|
|
|
|
 |
N.J.Kershaw,
M.E.Caines,
M.C.Sleeman,
and
C.J.Schofield
(2005).
The enzymology of clavam and carbapenem biosynthesis.
|
| |
Chem Commun (Camb),
(),
4251-4263.
|
 |
|
|
|
|
 |
S.Shimada,
Y.T.Inoue,
and
M.Sakuta
(2005).
Anthocyanidin synthase in non-anthocyanin-producing Caryophyllales species.
|
| |
Plant J,
44,
950-959.
|
 |
|
|
|
|
 |
T.Hansen,
B.Schlichting,
M.Felgendreher,
and
P.Schönheit
(2005).
Cupin-type phosphoglucose isomerases (Cupin-PGIs) constitute a novel metal-dependent PGI family representing a convergent line of PGI evolution.
|
| |
J Bacteriol,
187,
1621-1631.
|
 |
|
|
|
|
 |
X.B.Wu,
K.Q.Fan,
Q.H.Wang,
and
K.Q.Yang
(2005).
C-terminus mutations of Acremonium chrysogenum deacetoxy/deacetylcephalosporin C synthase with improved activity toward penicillin analogs.
|
| |
FEMS Microbiol Lett,
246,
103-110.
|
 |
|
|
|
|
 |
L.L.Videau,
W.B.Arendall,
and
J.S.Richardson
(2004).
The cis-Pro touch-turn: a rare motif preferred at functional sites.
|
| |
Proteins,
56,
298-309.
|
 |
|
|
|
|
 |
Z.Zhang,
J.S.Ren,
I.J.Clifton,
and
C.J.Schofield
(2004).
Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase--the ethylene-forming enzyme.
|
| |
Chem Biol,
11,
1383-1394.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.L.Wei,
Y.B.Yang,
W.C.Wang,
W.C.Liu,
J.S.Hsu,
and
Y.C.Tsai
(2003).
Engineering Streptomyces clavuligerus deacetoxycephalosporin C synthase for optimal ring expansion activity toward penicillin G.
|
| |
Appl Environ Microbiol,
69,
2306-2312.
|
 |
|
|
|
|
 |
C.Lee,
S.J.Kim,
D.G.Jeong,
S.M.Lee,
and
S.E.Ryu
(2003).
Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau.
|
| |
J Biol Chem,
278,
7558-7563.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.K.Kang,
J.Jeong,
S.K.Drake,
N.B.Wehr,
T.A.Rouault,
and
R.L.Levine
(2003).
Iron regulatory protein 2 as iron sensor. Iron-dependent oxidative modification of cysteine.
|
| |
J Biol Chem,
278,
14857-14864.
|
 |
|
|
|
|
 |
D.Lando,
J.J.Gorman,
M.L.Whitelaw,
and
D.J.Peet
(2003).
Oxygen-dependent regulation of hypoxia-inducible factors by prolyl and asparaginyl hydroxylation.
|
| |
Eur J Biochem,
270,
781-790.
|
 |
|
|
|
|
 |
E.I.Solomon,
A.Decker,
and
N.Lehnert
(2003).
Non-heme iron enzymes: contrasts to heme catalysis.
|
| |
Proc Natl Acad Sci U S A,
100,
3589-3594.
|
 |
|
|
|
|
 |
P.Liu,
A.Liu,
F.Yan,
M.D.Wolfe,
J.D.Lipscomb,
and
H.W.Liu
(2003).
Biochemical and spectroscopic studies on (S)-2-hydroxypropylphosphonic acid epoxidase: a novel mononuclear non-heme iron enzyme.
|
| |
Biochemistry,
42,
11577-11586.
|
 |
|
|
|
|
 |
R.W.Welford,
I.Schlemminger,
L.A.McNeill,
K.S.Hewitson,
and
C.J.Schofield
(2003).
The selectivity and inhibition of AlkB.
|
| |
J Biol Chem,
278,
10157-10161.
|
 |
|
|
|
|
 |
A.K.White,
and
W.W.Metcalf
(2002).
Isolation and biochemical characterization of hypophosphite/2-oxoglutarate dioxygenase. A novel phosphorus-oxidizing enzyme from Psuedomonas stutzeri WM88.
|
| |
J Biol Chem,
277,
38262-38271.
|
 |
|
|
|
|
 |
A.Serretti
(2002).
Lithium long-term treatment in mood disorders: clinical and genetic predictors.
|
| |
Pharmacogenomics,
3,
117-129.
|
 |
|
|
|
|
 |
C.E.Dann,
R.K.Bruick,
and
J.Deisenhofer
(2002).
Structure of factor-inhibiting hypoxia-inducible factor 1: An asparaginyl hydroxylase involved in the hypoxic response pathway.
|
| |
Proc Natl Acad Sci U S A,
99,
15351-15356.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.M.Hensgens,
E.A.Kroezinga,
B.A.van Montfort,
J.M.van der Laan,
J.D.Sutherland,
and
B.W.Dijkstra
(2002).
Purification, crystallization and preliminary X-ray diffraction of Cys103Ala acyl coenzyme A: isopenicillin N acyltransferase from Penicillium chrysogenum.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
716-718.
|
 |
|
|
|
|
 |
F.Wellmann,
R.Lukacin,
T.Moriguchi,
L.Britsch,
E.Schiltz,
and
U.Matern
(2002).
Functional expression and mutational analysis of flavonol synthase from Citrus unshiu.
|
| |
Eur J Biochem,
269,
4134-4142.
|
 |
|
|
|
|
 |
G.C.Ferreira,
R.Franco,
A.Mangravita,
and
G.N.George
(2002).
Unraveling the substrate-metal binding site of ferrochelatase: an X-ray absorption spectroscopic study.
|
| |
Biochemistry,
41,
4809-4818.
|
 |
|
|
|
|
 |
M.R.Chance,
A.R.Bresnick,
S.K.Burley,
J.S.Jiang,
C.D.Lima,
A.Sali,
S.C.Almo,
J.B.Bonanno,
J.A.Buglino,
S.Boulton,
H.Chen,
N.Eswar,
G.He,
R.Huang,
V.Ilyin,
L.McMahan,
U.Pieper,
S.Ray,
M.Vidal,
and
L.K.Wang
(2002).
Structural genomics: a pipeline for providing structures for the biologist.
|
| |
Protein Sci,
11,
723-738.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.Spielmeyer,
M.H.Ellis,
and
P.M.Chandler
(2002).
Semidwarf (sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene.
|
| |
Proc Natl Acad Sci U S A,
99,
9043-9048.
|
 |
|
|
|
|
 |
I.J.Clifton,
L.C.Hsueh,
J.E.Baldwin,
K.Harlos,
and
C.J.Schofield
(2001).
Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases.
|
| |
Eur J Biochem,
268,
6625-6636.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
J.J.Turnbull,
A.G.Prescott,
C.J.Schofield,
and
R.C.Wilmouth
(2001).
Purification, crystallization and preliminary X-ray diffraction of anthocyanidin synthase from Arabidopsis thaliana.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
425-427.
|
 |
|
|
|
|
 |
J.M.Ogle,
I.J.Clifton,
P.J.Rutledge,
J.M.Elkins,
N.I.Burzlaff,
R.M.Adlington,
P.L.Roach,
and
J.E.Baldwin
(2001).
Alternative oxidation by isopenicillin N synthase observed by X-ray diffraction.
|
| |
Chem Biol,
8,
1231-1237.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Loke,
and
T.S.Sim
(2001).
Mutational analysis of conserved glycines 42 and 256 in Cephalosporium acremonium isopenicillin N synthase.
|
| |
Can J Microbiol,
47,
961-964.
|
 |
|
|
|
|
 |
B.O.Bachmann,
and
C.A.Townsend
(2000).
Kinetic mechanism of the beta-lactam synthetase of Streptomyces clavuligerus.
|
| |
Biochemistry,
39,
11187-11193.
|
 |
|
|
|
|
 |
D.A.Hogan,
S.R.Smith,
E.A.Saari,
J.McCracken,
and
R.P.Hausinger
(2000).
Site-directed mutagenesis of 2,4-dichlorophenoxyacetic acid/alpha-ketoglutarate dioxygenase. Identification of residues involved in metallocenter formation and substrate binding.
|
| |
J Biol Chem,
275,
12400-12409.
|
 |
|
|
|
|
 |
R.Lukacin,
I.Gröning,
U.Pieper,
and
U.Matern
(2000).
Site-directed mutagenesis of the active site serine290 in flavanone 3beta-hydroxylase from Petunia hybrida.
|
| |
Eur J Biochem,
267,
853-860.
|
 |
|
|
|
|
 |
W.A.Schenk
(2000).
Isopenicillin N Synthase: An Enzyme at Work.
|
| |
Angew Chem Int Ed Engl,
39,
3409-3411.
|
 |
|
|
|
|
 |
A.M.Rocklin,
D.L.Tierney,
V.Kofman,
N.M.Brunhuber,
B.M.Hoffman,
R.E.Christoffersen,
N.O.Reich,
J.D.Lipscomb,
and
L.Que
(1999).
Role of the nonheme Fe(II) center in the biosynthesis of the plant hormone ethylene.
|
| |
Proc Natl Acad Sci U S A,
96,
7905-7909.
|
 |
|
|
|
|
 |
C.J.Schofield,
and
Z.Zhang
(1999).
Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes.
|
| |
Curr Opin Struct Biol,
9,
722-731.
|
 |
|
|
|
|
 |
E.L.Hegg,
A.K.Whiting,
R.E.Saari,
J.McCracken,
R.P.Hausinger,
and
L.Que
(1999).
Herbicide-degrading alpha-keto acid-dependent enzyme TfdA: metal coordination environment and mechanistic insights.
|
| |
Biochemistry,
38,
16714-16726.
|
 |
|
|
|
|
 |
M.J.Ryle,
R.Padmakumar,
and
R.P.Hausinger
(1999).
Stopped-flow kinetic analysis of Escherichia coli taurine/alpha-ketoglutarate dioxygenase: interactions with alpha-ketoglutarate, taurine, and oxygen.
|
| |
Biochemistry,
38,
15278-15286.
|
 |
|
|
|
|
 |
P.Loke,
and
T.Sim
(1999).
Site-directed mutagenesis of arginine-89 supports the role of its guanidino side-chain in substrate binding by Cephalosporium acremonium isopenicillin N synthase.
|
| |
FEMS Microbiol Lett,
179,
423-429.
|
 |
|
|
|
|
 |
A.A.Brakhage
(1998).
Molecular regulation of beta-lactam biosynthesis in filamentous fungi.
|
| |
Microbiol Mol Biol Rev,
62,
547-585.
|
 |
|
|
|
|
 |
A.M.Reeve,
S.D.Breazeale,
and
C.A.Townsend
(1998).
Purification, characterization, and cloning of an S-adenosylmethionine-dependent 3-amino-3-carboxypropyltransferase in nocardicin biosynthesis.
|
| |
J Biol Chem,
273,
30695-30703.
|
 |
|
|
|
|
 |
B.Kauppi,
K.Lee,
E.Carredano,
R.E.Parales,
D.T.Gibson,
H.Eklund,
and
S.Ramaswamy
(1998).
Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase.
|
| |
Structure,
6,
571-586.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.O.Bachmann,
R.Li,
and
C.A.Townsend
(1998).
beta-Lactam synthetase: a new biosynthetic enzyme.
|
| |
Proc Natl Acad Sci U S A,
95,
9082-9086.
|
 |
|
|
|
|
 |
C.J.Rowe,
C.P.Shorrock,
T.D.Claridge,
and
J.D.Sutherland
(1998).
Analysis of the conversion of delta-(L-alpha-aminoadipoyl)-L-cysteinyl-D-alpha-aminobutyrate by active-site mutants of Aspergillus nidulans isopenicillin N synthase.
|
| |
Chem Biol,
5,
229-239.
|
 |
|
|
|
|
 |
J.C.Milne,
A.C.Eliot,
N.L.Kelleher,
and
C.T.Walsh
(1998).
ATP/GTP hydrolysis is required for oxazole and thiazole biosynthesis in the peptide antibiotic microcin B17.
|
| |
Biochemistry,
37,
13250-13261.
|
 |
|
|
|
|
 |
P.Loke,
and
T.S.Sim
(1998).
Mutational evidence for the role of serine-283 in Cephalosporium acremonium isopenicillin N synthase.
|
| |
FEMS Microbiol Lett,
165,
353-356.
|
 |
|
|
|
|
 |
R.W.Frazee,
A.M.Orville,
K.B.Dolbeare,
H.Yu,
D.H.Ohlendorf,
and
J.D.Lipscomb
(1998).
The axial tyrosinate Fe3+ ligand in protocatechuate 3,4-dioxygenase influences substrate binding and product release: evidence for new reaction cycle intermediates.
|
| |
Biochemistry,
37,
2131-2144.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.J.Lange,
and
L.Que
(1998).
Oxygen activating nonheme iron enzymes.
|
| |
Curr Opin Chem Biol,
2,
159-172.
|
 |
|
|
|
|
 |
C.A.Townsend
(1997).
Structural studies of natural product biosynthetic proteins.
|
| |
Chem Biol,
4,
721-730.
|
 |
|
|
|
|
 |
C.J.Schofield,
J.E.Baldwin,
M.F.Byford,
I.Clifton,
J.Hajdu,
C.Hensgens,
and
P.Roach
(1997).
Proteins of the penicillin biosynthesis pathway.
|
| |
Curr Opin Struct Biol,
7,
857-864.
|
 |
|
|
|
|
 |
E.L.Hegg,
and
L.Que
(1997).
The 2-His-1-carboxylate facial triad--an emerging structural motif in mononuclear non-heme iron(II) enzymes.
|
| |
Eur J Biochem,
250,
625-629.
|
 |
|
|
|
|
 |
P.Loke,
J.Sim,
and
T.S.Sim
(1997).
Functional analysis of a conserved aspartate D218 in Cephalosporium acremonium isopenicillin N synthase.
|
| |
FEMS Microbiol Lett,
157,
137-140.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

