 |
PDBsum entry 6zg0

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 1 more)
143 a.a.
(+ 1 more)
143 a.a.
|
 |

|

|

|

|

|

|

|
 157 a.a.
157 a.a.
|
 |

|

|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Sarm1 sam1-2 domains
|
|
Structure:
|
 |
NAD(+) hydrolase sarm1. Chain: a, b, c, d, e, f, g, h. Synonym: hsarm1,NADP(+) hydrolase sarm1,sterile alpha and armadillo repeat protein,sterile alpha and tir motif-containing protein 1, sterile alpha motif domain-containing protein 2,sam domain-containing protein 2,tir-1 homolog,hstir. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: sarm1, kiaa0524, samd2, sarm. Expressed in: homo sapiens. Expression_system_taxid: 9606. Expression_system_cell_line: hek293f
|
|
Authors:
|
 |
M.Sporny,J.Guez-Haddad,T.Khazma,A.Yaron,M.Dessau,C.Mim,M.N.Isupov, R.Zalk,M.Hons,Y.Opatowsky
|
|
Key ref:
|
 |
m.sporny
et al.
(2020).
The structural basis for SARM1 inhibition and activation under energetic stress.
Elife,
9,
.
PubMed id:
|
 |
|
Date:
|
 |
|
18-Jun-20
|
Release date:
|
11-Nov-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.3.2.2.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B, C, D, E, F, G, H:
E.C.3.2.2.6
- ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
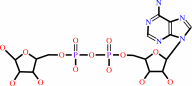 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Elife
9:
(2020)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis for SARM1 inhibition and activation under energetic stress.
|
|
m.sporny,
j.guez-haddad,
t.khazma,
a.yaron,
m.dessau,
c.mim.n.isupov,
r.zalk,
m.hons,
y.opatowsky.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

