 |
PDBsum entry 6cbh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Cytokine
|
 |
|
Title:
|
 |
Macrophage migration inhibitory factor in complex with a pyrazole inhibitor (8m)
|
|
Structure:
|
 |
Macrophage migration inhibitory factor. Chain: a, b, c. Synonym: mif,glycosylation-inhibiting factor,gif,l-dopachrome isomerase,l-dopachrome tautomerase,phenylpyruvate tautomerase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: mif, glif, mmif. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.177
|
R-free:
|
0.214
|
|
|
Authors:
|
 |
M.J.Robertson,S.G.Krimmer,W.L.Jorgensen
|
|
Key ref:
|
 |
V.Trivedi-Parmar
et al.
(2018).
Optimization of Pyrazoles as Phenol Surrogates to Yield Potent Inhibitors of Macrophage Migration Inhibitory Factor.
ChemMedChem,
13,
1092-1097.
PubMed id:
|
 |
|
Date:
|
 |
|
02-Feb-18
|
Release date:
|
04-Apr-18
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P14174
(MIF_HUMAN) -
Macrophage migration inhibitory factor from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
115 a.a.
114 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.3.2.1
- phenylpyruvate tautomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-phenylpyruvate = enol-phenylpyruvate
|
 |
 |
 |
 |
 |
3-phenylpyruvate
|
=
|
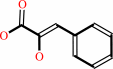 enol-phenylpyruvate
enol-phenylpyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.3.3.12
- L-dopachrome isomerase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-dopachrome = 5,6-dihydroxyindole-2-carboxylate
|
 |
 |
 |
 |
 |
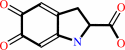 L-dopachrome
L-dopachrome
|
=
|
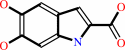 5,6-dihydroxyindole-2-carboxylate
5,6-dihydroxyindole-2-carboxylate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
ChemMedChem
13:1092-1097
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Optimization of Pyrazoles as Phenol Surrogates to Yield Potent Inhibitors of Macrophage Migration Inhibitory Factor.
|
|
V.Trivedi-Parmar,
M.J.Robertson,
J.A.Cisneros,
S.G.Krimmer,
W.L.Jorgensen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Macrophage migration inhibitory factor (MIF) is a proinflammatory cytokine that
is implicated in the regulation of inflammation, cell proliferation, and
neurological disorders. MIF is also an enzyme that functions as a keto-enol
tautomerase. Most potent MIF tautomerase inhibitors incorporate a phenol, which
hydrogen bonds to Asn97 in the active site. Starting from a 113-μm docking hit,
we report results of structure-based and computer-aided design that have
provided substituted pyrazoles as phenol alternatives with potencies of
60-70 nm. Crystal structures of complexes of MIF with the pyrazoles highlight
the contributions of hydrogen bonding with Lys32 and Asn97, and aryl-aryl
interactions with Tyr36, Tyr95, and Phe113 to the binding.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

