 |
PDBsum entry 3log

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Crystal structure of mbti from mycobacterium tuberculosis
|
|
Structure:
|
 |
Isochorismate synthase/isochorismate-pyruvate lyase mbti. Chain: a, b, c, d. Synonym: salicylate synthase mbti, mycobactin synthetase protein i. Engineered: yes
|
|
Source:
|
 |
Mycobacterium tuberculosis. Organism_taxid: 1773. Gene: mbti, mt2454, rv2386c. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.73Å
|
R-factor:
|
0.183
|
R-free:
|
0.225
|
|
|
Authors:
|
 |
E.M.M.Bulloch,J.S.Lott,E.N.Baker,J.M.Johnston
|
|
Key ref:
|
 |
A.Manos-Turvey
et al.
(2010).
Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI).
Chemmedchem,
5,
1067-1079.
PubMed id:
|
 |
|
Date:
|
 |
|
03-Feb-10
|
Release date:
|
09-Feb-11
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WFX1
(MBTI_MYCTU) -
Salicylate synthase from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
450 a.a.
435 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.2.99.21
- isochorismate lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isochorismate = salicylate + pyruvate
|
 |
 |
 |
 |
 |
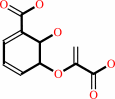 isochorismate
isochorismate
|
=
|
salicylate
Bound ligand (Het Group name = )
matches with 55.56% similarity
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.5.4.4.2
- isochorismate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chorismate = isochorismate
|
 |
 |
 |
 |
 |
chorismate
Bound ligand (Het Group name = )
matches with 41.18% similarity
|
=
|
 isochorismate
isochorismate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.5.4.99.5
- chorismate mutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
chorismate = prephenate
|
 |
 |
 |
 |
 |
chorismate
Bound ligand (Het Group name = )
matches with 41.18% similarity
|
=
|
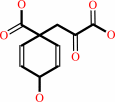 prephenate
prephenate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Chemmedchem
5:1067-1079
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI).
|
|
A.Manos-Turvey,
E.M.Bulloch,
P.J.Rutledge,
E.N.Baker,
J.S.Lott,
R.J.Payne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Mycobacterium tuberculosis salicylate synthase (MbtI), a member of the
chorismate-utilizing enzyme family, catalyses the first committed step in the
biosynthesis of the siderophore mycobactin T. This complex secondary metabolite
is essential for both virulence and survival of M. tuberculosis, the etiological
agent of tuberculosis (TB). It is therefore anticipated that inhibitors of this
enzyme may serve as TB therapies with a novel mode of action. Herein we describe
the first inhibition study of M. tuberculosis MbtI using a library of
functionalized benzoate-based inhibitors designed to mimic the substrate
(chorismate) and intermediate (isochorismate) of the MbtI-catalyzed reaction.
The most potent inhibitors prepared were those designed to mimic the enzyme
intermediate, isochorismate. These compounds, based on a 2,3-dihydroxybenzoate
scaffold, proved to be low-micromolar inhibitors of MbtI. The most potent
inhibitors in this series possessed hydrophobic enol ether side chains at C3 in
place of the enol-pyruvyl side chain found in chorismate and isochorismate.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Vasan,
J.Neres,
J.Williams,
D.J.Wilson,
A.M.Teitelbaum,
R.P.Remmel,
and
C.C.Aldrich
(2010).
Inhibitors of the salicylate synthase (MbtI) from Mycobacterium tuberculosis discovered by high-throughput screening.
|
| |
ChemMedChem,
5,
2079-2087.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

