 |
PDBsum entry 2qw8

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2qw8
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.318
- eugenol synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
eugenol + a carboxylate + NADP+ = a coniferyl ester + NADPH
|
 |
 |
 |
 |
 |
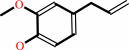 eugenol
eugenol
|
+
|
 carboxylate
carboxylate
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
coniferyl ester
|
+
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Plos One
2:e993
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and reaction mechanism of basil eugenol synthase.
|
|
G.V.Louie,
T.J.Baiga,
M.E.Bowman,
T.Koeduka,
J.H.Taylor,
S.M.Spassova,
E.Pichersky,
J.P.Noel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phenylpropenes, a large group of plant volatile compounds that serve in multiple
roles in defense and pollinator attraction, contain a propenyl side chain.
Eugenol synthase (EGS) catalyzes the reductive displacement of acetate from the
propenyl side chain of the substrate coniferyl acetate to produce the
allyl-phenylpropene eugenol. We report here the structure determination of EGS
from basil (Ocimum basilicum) by protein x-ray crystallography. EGS is
structurally related to the short-chain dehydrogenase/reductases (SDRs), and in
particular, enzymes in the isoflavone-reductase-like subfamily. The structure of
a ternary complex of EGS bound to the cofactor NADP(H) and a mixed competitive
inhibitor EMDF ((7S,8S)-ethyl (7,8-methylene)-dihydroferulate) provides a
detailed view of the binding interactions within the EGS active site and a
starting point for mutagenic examination of the unusual reductive mechanism of
EGS. The key interactions between EMDF and the EGS-holoenzyme include stacking
of the phenyl ring of EMDF against the cofactor's nicotinamide ring and a
water-mediated hydrogen-bonding interaction between the EMDF 4-hydroxy group and
the side-chain amino moiety of a conserved lysine residue, Lys132. The C4 carbon
of nicotinamide resides immediately adjacent to the site of hydride addition,
the C7 carbon of cinnamyl acetate substrates. The inhibitor-bound EGS structure
suggests a two-step reaction mechanism involving the formation of a
quinone-methide prior to reduction. The formation of this intermediate is
promoted by a hydrogen-bonding network that favors deprotonation of the
substrate's 4-hydroxyl group and disfavors binding of the acetate moiety, akin
to a push-pull catalytic mechanism. Notably, the catalytic involvement in EGS of
the conserved Lys132 in preparing the phenolic substrate for quinone methide
formation through the proton-relay network appears to be an adaptation of the
analogous role in hydrogen bonding played by the equivalent lysine residue in
other enzymes of the SDR family.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Gargouri,
J.Chaudière,
C.Manigand,
C.Maugé,
K.Bathany,
J.M.Schmitter,
and
B.Gallois
(2010).
The epimerase activity of anthocyanidin reductase from Vitis vinifera and its regiospecific hydride transfers.
|
| |
Biol Chem,
391,
219-227.
|
 |
|
|
|
|
 |
T.Vogt
(2010).
Phenylpropanoid biosynthesis.
|
| |
Mol Plant,
3,
2.
|
 |
|
|
|
|
 |
M.Kajikawa,
N.Hirai,
and
T.Hashimoto
(2009).
A PIP-family protein is required for biosynthesis of tobacco alkaloids.
|
| |
Plant Mol Biol,
69,
287-298.
|
 |
|
|
|
|
 |
L.B.Davin,
M.Jourdes,
A.M.Patten,
K.W.Kim,
D.G.Vassão,
and
N.G.Lewis
(2008).
Dissection of lignin macromolecular configuration and assembly: Comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis.
|
| |
Nat Prod Rep,
25,
1015-1090.
|
 |
|
|
|
|
 |
T.Koeduka,
G.V.Louie,
I.Orlova,
C.M.Kish,
M.Ibdah,
C.G.Wilkerson,
M.E.Bowman,
T.J.Baiga,
J.P.Noel,
N.Dudareva,
and
E.Pichersky
(2008).
The multiple phenylpropene synthases in both Clarkia breweri and Petunia hybrida represent two distinct protein lineages.
|
| |
Plant J,
54,
362-374.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
U.Bayindir,
A.W.Alfermann,
and
E.Fuss
(2008).
Hinokinin biosynthesis in Linum corymbulosum Reichenb.
|
| |
Plant J,
55,
810-820.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

