 |
PDBsum entry 2jch

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Drug-binding protein
|
PDB id
|
|

|

|
2jch
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.99.28
- peptidoglycan glycosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)](n)- di-trans,octa-cis-undecaprenyl diphosphate + beta-D-GlcNAc-(1->4)- Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-di-trans,octa- cis-undecaprenyl diphosphate = [GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D- Glu-L-Lys-D-Ala-D-Ala)](n+1)-di-trans,octa-cis-undecaprenyl diphosphate + di-trans,octa-cis-undecaprenyl diphosphate + H+
|
 |
 |
 |
 |
 |
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)](n)- di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
beta-D-GlcNAc-(1->4)- Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-di-trans,octa- cis-undecaprenyl diphosphate
|
=
|
[GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D- Glu-L-Lys-D-Ala-D-Ala)](n+1)-di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
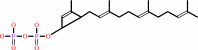 di-trans,octa-cis-undecaprenyl diphosphate
di-trans,octa-cis-undecaprenyl diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Chem Biol
3:565-569
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and mechanistic basis of penicillin-binding protein inhibition by lactivicins.
|
|
P.Macheboeuf,
D.S.Fischer,
T.Brown,
A.Zervosen,
A.Luxen,
B.Joris,
A.Dessen,
C.J.Schofield.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Beta-lactam antibiotics, including penicillins and cephalosporins, inhibit
penicillin-binding proteins (PBPs), which are essential for bacterial cell wall
biogenesis. Pathogenic bacteria have evolved efficient antibiotic resistance
mechanisms that, in Gram-positive bacteria, include mutations to PBPs that
enable them to avoid beta-lactam inhibition. Lactivicin (LTV; 1) contains
separate cycloserine and gamma-lactone rings and is the only known natural PBP
inhibitor that does not contain a beta-lactam. Here we show that LTV and a more
potent analog, phenoxyacetyl-LTV (PLTV; 2), are active against clinically
isolated, penicillin-resistant Streptococcus pneumoniae strains.
Crystallographic analyses of S. pneumoniae PBP1b reveal that LTV and PLTV
inhibition involves opening of both monocyclic cycloserine and gamma-lactone
rings. In PBP1b complexes, the ring-derived atoms from LTV and PLTV show a
notable structural convergence with those derived from a complexed cephalosporin
(cefotaxime; 3). The structures imply that derivatives of LTV will be useful in
the search for new antibiotics with activity against beta-lactam-resistant
bacteria.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
(a) Structures of  -lactam
antibiotics. The conserved -lactam
antibiotics. The conserved  -lactam
ring is highlighted in green. (b) Structures of epimeric LTV and
PLTV. (c) Outline mechanism for -lactam
ring is highlighted in green. (b) Structures of epimeric LTV and
PLTV. (c) Outline mechanism for  -lactams
showing formation of the hydrolytically stable acyl-enzyme
complex (for cefotaxime: R, aminothiazolemethoxyoxime; X,
OCOCH[3]). (d) Proposed mechanism of PBP acylation by LTV and
PLTV, involving formation of a stable acyl-enzyme complex whose
structure is closely analogous to that formed by cephalosporins. -lactams
showing formation of the hydrolytically stable acyl-enzyme
complex (for cefotaxime: R, aminothiazolemethoxyoxime; X,
OCOCH[3]). (d) Proposed mechanism of PBP acylation by LTV and
PLTV, involving formation of a stable acyl-enzyme complex whose
structure is closely analogous to that formed by cephalosporins.
|
 |
Figure 3.
(a) F[o] – F[c] map (green) contoured at 2.4  ,
generated before inclusion of LTV in the model. (b) F[o] –
F[c] map (green), contoured at 2.4 ,
generated before inclusion of LTV in the model. (b) F[o] –
F[c] map (green), contoured at 2.4  ,
for the PLTV molecule. Selected active site residues are shown
as sticks; LTV and PLTV backbones are shown in blue. Notably,
ester link is present between the Ser460 side chain and the
carbonyl of the LTV/PLTV cycloserine ring, and both cycloserine
and lactone rings of the inhibitors are open. Figure was
prepared using PyMOL (http://pymol.sourceforge.net/). ,
for the PLTV molecule. Selected active site residues are shown
as sticks; LTV and PLTV backbones are shown in blue. Notably,
ester link is present between the Ser460 side chain and the
carbonyl of the LTV/PLTV cycloserine ring, and both cycloserine
and lactone rings of the inhibitors are open. Figure was
prepared using PyMOL (http://pymol.sourceforge.net/).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Chem Biol
(2007,
3,
565-569)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.F.Kluge,
and
R.C.Petter
(2010).
Acylating drugs: redesigning natural covalent inhibitors.
|
| |
Curr Opin Chem Biol,
14,
421-427.
|
 |
|
|
|
|
 |
J.Lowther,
B.A.Yard,
K.A.Johnson,
L.G.Carter,
V.T.Bhat,
M.C.Raman,
D.J.Clarke,
B.Ramakers,
S.A.McMahon,
J.H.Naismith,
and
D.J.Campopiano
(2010).
Inhibition of the PLP-dependent enzyme serine palmitoyltransferase by cycloserine: evidence for a novel decarboxylative mechanism of inactivation.
|
| |
Mol Biosyst,
6,
1682-1693.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Lemaire,
C.Fuda,
F.Van Bambeke,
P.M.Tulkens,
and
S.Mobashery
(2008).
Restoration of susceptibility of methicillin-resistant Staphylococcus aureus to beta-lactam antibiotics by acidic pH: role of penicillin-binding protein PBP 2a.
|
| |
J Biol Chem,
283,
12769-12776.
|
 |
|
|
|
|
 |
U.Holzgrabe
(2007).
[Lactivicin--an antibiotic against penicillin-resistant pneumococci]
|
| |
Pharm Unserer Zeit,
36,
421-422.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

