 |
PDBsum entry 2inx

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.3.1
- steroid Delta-isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 3-oxo-Delta5-steroid = a 3-oxo-Delta4-steroid
|
 |
 |
 |
 |
 |
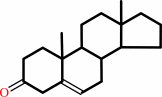 3-oxo-Delta(5)-steroid
3-oxo-Delta(5)-steroid
|
=
|
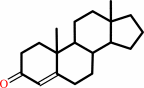 3-oxo-Delta(4)-steroid
3-oxo-Delta(4)-steroid
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Am Chem Soc
130:13696-13708
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
|
|
P.A.Sigala,
D.A.Kraut,
J.M.Caaveiro,
B.Pybus,
E.A.Ruben,
D.Ringe,
G.A.Petsko,
D.Herschlag.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Enzymes are classically proposed to accelerate reactions by binding substrates
within active-site environments that are structurally preorganized to optimize
binding interactions with reaction transition states rather than ground states.
This is a remarkably formidable task considering the limited 0.1-1 A scale of
most substrate rearrangements. The flexibility of active-site functional groups
along the coordinate of substrate rearrangement, the distance scale on which
enzymes can distinguish structural rearrangement, and the energetic significance
of discrimination on that scale remain open questions that are fundamental to a
basic physical understanding of enzyme active sites and catalysis. We bring
together 1.2-1.5 A resolution X-ray crystallography, (1)H and (19)F NMR
spectroscopy, quantum mechanical calculations, and transition-state analogue
binding measurements to test the distance scale on which noncovalent forces can
constrain the structural relaxation or translation of side chains and ligands
along a specific coordinate and the energetic consequences of such geometric
constraints within the active site of bacterial ketosteroid isomerase (KSI). Our
results strongly suggest that packing and binding interactions within the KSI
active site can constrain local side-chain reorientation and prevent hydrogen
bond shortening by 0.1 A or less. Further, this constraint has substantial
energetic effects on ligand binding and stabilization of negative charge within
the oxyanion hole. These results provide evidence that subtle geometric effects,
indistinguishable in most X-ray crystallographic structures, can have
significant energetic consequences and highlight the importance of using
synergistic experimental approaches to dissect enzyme function.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Armenta-Medina,
E.Pérez-Rueda,
and
L.Segovia
(2011).
Identification of functional motions in the adenylate kinase (ADK) protein family by computational hybrid approaches.
|
| |
Proteins,
79,
1662-1671.
|
 |
|
|
|
|
 |
S.H.Ackerman,
and
D.L.Gatti
(2011).
The contribution of coevolving residues to the stability of KDO8P synthase.
|
| |
PLoS One,
6,
e17459.
|
 |
|
|
|
|
 |
D.A.Kraut,
P.A.Sigala,
T.D.Fenn,
and
D.Herschlag
(2010).
Dissecting the paradoxical effects of hydrogen bond mutations in the ketosteroid isomerase oxyanion hole.
|
| |
Proc Natl Acad Sci U S A,
107,
1960-1965.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.C.Brookes
(2010).
Science is perception: what can our sense of smell tell us about ourselves and the world around us?
|
| |
Philos Transact A Math Phys Eng Sci,
368,
3491-3502.
|
 |
|
|
|
|
 |
K.L.Peña,
S.E.Castel,
C.de Araujo,
G.S.Espie,
and
M.S.Kimber
(2010).
Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, CcmM.
|
| |
Proc Natl Acad Sci U S A,
107,
2455-2460.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.C.Kamerlin,
and
A.Warshel
(2010).
At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis?
|
| |
Proteins,
78,
1339-1375.
|
 |
|
|
|
|
 |
S.C.Kamerlin,
P.K.Sharma,
Z.T.Chu,
and
A.Warshel
(2010).
Ketosteroid isomerase provides further support for the idea that enzymes work by electrostatic preorganization.
|
| |
Proc Natl Acad Sci U S A,
107,
4075-4080.
|
 |
|
|
|
|
 |
D.A.Kraut,
M.J.Churchill,
P.E.Dawson,
and
D.Herschlag
(2009).
Evaluating the potential for halogen bonding in the oxyanion hole of ketosteroid isomerase using unnatural amino acid mutagenesis.
|
| |
ACS Chem Biol,
4,
269-273.
|
 |
|
|
|
|
 |
D.K.Chakravorty,
A.V.Soudackov,
and
S.Hammes-Schiffer
(2009).
Hybrid quantum/classical molecular dynamics simulations of the proton transfer reactions catalyzed by ketosteroid isomerase: analysis of hydrogen bonding, conformational motions, and electrostatics.
|
| |
Biochemistry,
48,
10608-10619.
|
 |
|
|
|
|
 |
E.Di Cera
(2009).
Serine proteases.
|
| |
IUBMB Life,
61,
510-515.
|
 |
|
|
|
|
 |
J.Damborsky,
and
J.Brezovsky
(2009).
Computational tools for designing and engineering biocatalysts.
|
| |
Curr Opin Chem Biol,
13,
26-34.
|
 |
|
|
|
|
 |
J.P.Schwans,
D.A.Kraut,
and
D.Herschlag
(2009).
Determining the catalytic role of remote substrate binding interactions in ketosteroid isomerase.
|
| |
Proc Natl Acad Sci U S A,
106,
14271-14275.
|
 |
|
|
|
|
 |
P.A.Sigala,
M.A.Tsuchida,
and
D.Herschlag
(2009).
Hydrogen bond dynamics in the active site of photoactive yellow protein.
|
| |
Proc Natl Acad Sci U S A,
106,
9232-9237.
|
 |
|
|
|
|
 |
A.J.Kirby,
and
F.Hollfelder
(2008).
Biochemistry: Enzymes under the nanoscope.
|
| |
Nature,
456,
45-47.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

