 |
PDBsum entry 2i3c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of an aspartoacylase from homo sapiens
|
|
Structure:
|
 |
Aspartoacylase. Chain: a, b. Synonym: aminoacylase-2, acy-2. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: aspa, acy2, asp. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.197
|
R-free:
|
0.243
|
|
|
Authors:
|
 |
E.Bitto,G.E.Wesenberg,G.N.Phillips Jr.,J.G.Mccoy,C.A.Bingman,Center For Eukaryotic Structural Genomics (Cesg)
|
Key ref:

|
 |
E.Bitto
et al.
(2007).
Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc Natl Acad Sci U S A,
104,
456-461.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
17-Aug-06
|
Release date:
|
29-Aug-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P45381
(ACY2_HUMAN) -
Aspartoacylase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
313 a.a.
302 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.15
- aspartoacylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acyl-L-aspartate + H2O = a carboxylate + L-aspartate
|
 |
 |
 |
 |
 |
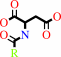 N-acyl-L-aspartate
N-acyl-L-aspartate
|
+
|
H2O
|
=
|
 carboxylate
carboxylate
|
+
|
 L-aspartate
L-aspartate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
104:456-461
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
|
|
E.Bitto,
C.A.Bingman,
G.E.Wesenberg,
J.G.McCoy,
G.N.Phillips.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aspartoacylase catalyzes hydrolysis of N-acetyl-l-aspartate to aspartate and
acetate in the vertebrate brain. Deficiency in this activity leads to spongiform
degeneration of the white matter of the brain and is the established cause of
Canavan disease, a fatal progressive leukodystrophy affecting young children. We
present crystal structures of recombinant human and rat aspartoacylase refined
to 2.8- and 1.8-A resolution, respectively. The structures revealed that the
N-terminal domain of aspartoacylase adopts a protein fold similar to that of
zinc-dependent hydrolases related to carboxypeptidases A. The catalytic site of
aspartoacylase shows close structural similarity to those of carboxypeptidases
despite only 10-13% sequence identity between these proteins. About 100
C-terminal residues of aspartoacylase form a globular domain with a two-stranded
beta-sheet linker that wraps around the N-terminal domain. The long channel
leading to the active site is formed by the interface of the N- and C-terminal
domains. The C-terminal domain is positioned in a way that prevents productive
binding of polypeptides in the active site. The structures revealed that
residues 158-164 may undergo a conformational change that results in opening and
partial closing of the channel entrance. We hypothesize that the catalytic
mechanism of aspartoacylase is closely analogous to that of carboxypeptidases.
We identify residues involved in zinc coordination, and propose which residues
may be involved in substrate binding and catalysis. The structures also provide
a structural framework necessary for understanding the deleterious effects of
many missense mutations of human aspartoacylase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Ribbon diagrams of the rASPA monomer and dimer. (A)
N-domain of rASPA is color-coded in cyan and red. C-domain is
color-coded in yellow and green. Residues His-21, Gly-22,
Glu-24, Asn-54, Arg-63, Asn-70, Arg-71, Phe-73, Asp-114,
His-116, and Glu-178 (blue sticks) are highly conserved in the
AstE-AspA family and delineate the active site. Zn^2+ is shown
as a pink sphere. (B) The rASPA dimer observed in the asymmetric
unit of the rASPA crystals is shown in ribbon representation.
Both the N-domain (red) and C-domain (green) of the rASPA
monomers are involved in formation of the dimer interface.
Residues His-21, Glu-24, and His-116 (blue sticks) coordinate
Zn^2+ (pink sphere).
|
 |
Figure 4.
Fig. 4. Proposed mechanism of action of ASPA.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Zano,
R.Malik,
S.Szucs,
R.Matalon,
and
R.E.Viola
(2011).
Modification of aspartoacylase for potential use in enzyme replacement therapy for the treatment of Canavan disease.
|
| |
Mol Genet Metab,
102,
176-180.
|
 |
|
|
|
|
 |
J.M.Hsieh,
K.Tsirulnikov,
M.R.Sawaya,
N.Magilnick,
N.Abuladze,
I.Kurtz,
J.Abramson,
and
A.Pushkin
(2010).
Structures of aminoacylase 3 in complex with acetylated substrates.
|
| |
Proc Natl Acad Sci U S A,
107,
17962-17967.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.H.Baslow
(2010).
Evidence that the tri-cellular metabolism of N-acetylaspartate functions as the brain's "operating system": how NAA metabolism supports meaningful intercellular frequency-encoded communications.
|
| |
Amino Acids,
39,
1139-1145.
|
 |
|
|
|
|
 |
A.Edwards
(2009).
Large-scale structural biology of the human proteome.
|
| |
Annu Rev Biochem,
78,
541-568.
|
 |
|
|
|
|
 |
K.Tsirulnikov,
N.Abuladze,
D.Newman,
S.Ryazantsev,
T.Wolak,
N.Magilnick,
M.C.Koag,
I.Kurtz,
and
A.Pushkin
(2009).
Mouse aminoacylase 3: a metalloenzyme activated by cobalt and nickel.
|
| |
Biochim Biophys Acta,
1794,
1049-1057.
|
 |
|
|
|
|
 |
M.H.Baslow,
and
D.N.Guilfoyle
(2009).
Are astrocytes the missing link between lack of brain aspartoacylase activity and the spongiform leukodystrophy in canavan disease?
|
| |
Neurochem Res,
34,
1523-1534.
|
 |
|
|
|
|
 |
R.E.Connon,
J.Geist,
J.Pfeiff,
A.V.Loguinov,
L.S.D'Abronzo,
H.Wintz,
C.D.Vulpe,
and
I.Werner
(2009).
Linking mechanistic and behavioral responses to sublethal esfenvalerate exposure in the endangered delta smelt; Hypomesus transpacificus (Fam. Osmeridae).
|
| |
BMC Genomics,
10,
608.
|
 |
|
|
|
|
 |
J.Le Coq,
A.Pavlovsky,
R.Malik,
R.Sanishvili,
C.Xu,
and
R.E.Viola
(2008).
Examination of the mechanism of human brain aspartoacylase through the binding of an intermediate analogue.
|
| |
Biochemistry,
47,
3484-3492.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Weigelt,
L.D.McBroom-Cerajewski,
M.Schapira,
Y.Zhao,
C.H.Arrowsmith,
and
C.H.Arrowmsmith
(2008).
Structural genomics and drug discovery: all in the family.
|
| |
Curr Opin Chem Biol,
12,
32-39.
|
 |
|
|
|
|
 |
N.Kaya,
F.Imtiaz,
D.Colak,
M.Al-Sayed,
A.Al-Odaib,
F.Al-Zahrani,
B.R.Al-Mubarak,
M.Al-Owain,
H.Al-Dhalaan,
A.Chedrawi,
Z.Al-Hassnan,
S.Coskun,
N.Sakati,
P.Ozand,
and
B.F.Meyer
(2008).
Genome-wide gene expression profiling and mutation analysis of Saudi patients with Canavan disease.
|
| |
Genet Med,
10,
675-684.
|
 |
|
|
|
|
 |
G.N.Phillips,
B.G.Fox,
J.L.Markley,
B.F.Volkman,
E.Bae,
E.Bitto,
C.A.Bingman,
R.O.Frederick,
J.G.McCoy,
B.L.Lytle,
B.S.Pierce,
J.Song,
and
S.N.Twigger
(2007).
Structures of proteins of biomedical interest from the Center for Eukaryotic Structural Genomics.
|
| |
J Struct Funct Genomics,
8,
73-84.
|
 |
|
|
|
|
 |
J.R.Hershfield,
N.Pattabiraman,
C.N.Madhavarao,
and
M.A.Namboodiri
(2007).
Mutational analysis of aspartoacylase: implications for Canavan disease.
|
| |
Brain Res,
1148,
1.
|
 |
|
|
|
|
 |
R.E.Viola
(2007).
The impact of structural biology on neurobiology.
|
| |
Proc Natl Acad Sci U S A,
104,
399-400.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

