 |
PDBsum entry 2ewb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.4.11.1
- leucyl aminopeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Release of an N-terminal amino acid, Xaa-|-Xbb-, in which Xaa is preferably Leu, but may be other amino acids including Pro although not Arg or Lys, and Xbb may be Pro.
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.4.11.5
- prolyl aminopeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Release of a N-terminal proline from a peptide.
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.4.13.23
- cysteinylglycine-S-conjugate dipeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an S-substituted L-cysteinylglycine + H2O = an S-substituted L-cysteine + glycine
|
 |
 |
 |
 |
 |
S-substituted L-cysteinylglycine
|
+
|
H2O
|
=
|
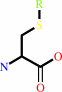 S-substituted L-cysteine
S-substituted L-cysteine
|
+
|
glycine
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
45:3226-3234
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Metal ion substitution in the catalytic site greatly affects the binding of sulfhydryl-containing compounds to leucyl aminopeptidase.
|
|
M.Cappiello,
V.Alterio,
P.Amodeo,
A.Del Corso,
A.Scaloni,
C.Pedone,
R.Moschini,
G.M.De Donatis,
G.De Simone,
U.Mura.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Bovine lens leucyl aminopeptidase (blLAP), a homohexameric metallopeptidase
preferring bulky and hydrophobic amino acids at the N-terminus of (di)peptides,
contains two Zn(2+) ions per subunit that are essential for catalytic activity.
They may be replaced by other divalent cations with different exchange kinetics.
The protein readily exchangeable site (site 1) can be occupied by Zn(2+),
Mn(2+), Mg(2+), or Co(2+), while the tight binding site (site 2) can be occupied
by Zn(2+) or Co(2+). We recently reported that introduction of Mn(2+) into site
1 generates a novel activity of blLAP toward CysGly [Cappiello, M., et al.
(2004) Biochem. J. 378, 35-44], which in contrast is not hydrolyzed by the
(Zn/Zn) enzyme. This finding, while disclosing a potential specific role for
blLAP in glutathione metabolism, raised a question about the features required
for molecules to be a substrate for the enzyme. To clarify the interaction of
the enzyme with sulfhydryl-containing derivatives, (Zn/Zn)- and (Mn/Zn)blLAP
forms were prepared and functional-structural studies were undertaken. Thus, a
kinetic analysis of various compounds with both enzyme forms was performed; the
crystal structure of (Zn/Zn)blLAP in complex with the peptidomimetic derivative
Zofenoprilat was determined, and a modeling study on the CysGly-(Zn/Zn)blLAP
complex was carried out. This combined approach provided insight into the
interaction of blLAP with sulfhydryl-containing derivatives, showing that the
metal exchange in site 1 modulates binding to these molecules that may result in
enzyme substrates or inhibitors, depending on the nature of the metal.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Pícha,
R.Liboska,
M.Buděšínský,
J.Jiráček,
M.Pawełczak,
and
A.Mucha
(2011).
Unusual activity pattern of leucine aminopeptidase inhibitors based on phosphorus containing derivatives of methionine and norleucine.
|
| |
J Enzyme Inhib Med Chem,
26,
155-161.
|
 |
|
|
|
|
 |
H.Ngyen,
M.Orlamuender,
D.Pretzel,
I.Agricola,
U.Sternberg,
and
S.Reissmann
(2008).
Transition metal complexes of a cyclic pseudo hexapeptide: synthesis, complex formation and catalytic activities.
|
| |
J Pept Sci,
14,
1010-1021.
|
 |
|
|
|
|
 |
L.Chu,
Y.Lai,
X.Xu,
S.Eddy,
S.Yang,
L.Song,
and
D.Kolodrubetz
(2008).
A 52-kDa leucyl aminopeptidase from treponema denticola is a cysteinylglycinase that mediates the second step of glutathione metabolism.
|
| |
J Biol Chem,
283,
19351-19358.
|
 |
|
|
|
|
 |
M.Matsui,
J.H.Fowler,
and
L.L.Walling
(2006).
Leucine aminopeptidases: diversity in structure and function.
|
| |
Biol Chem,
387,
1535-1544.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

