 |
PDBsum entry 1vj5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.76
- lipid-phosphate phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(9S,10S)-10-hydroxy-9-(phosphooxy)octadecanoate + H2O = (9S,10S)-9,10- dihydroxyoctadecanoate + phosphate
|
 |
 |
 |
 |
 |
(9S,10S)-10-hydroxy-9-(phosphooxy)octadecanoate
|
+
|
H2O
|
=
|
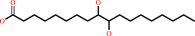 (9S,10S)-9,10- dihydroxyoctadecanoate
(9S,10S)-9,10- dihydroxyoctadecanoate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.3.2.10
- soluble epoxide hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an epoxide + H2O = an ethanediol
|
 |
 |
 |
 |
 |
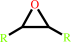 epoxide
epoxide
|
+
|
H2O
|
=
|
ethanediol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
43:4716-4723
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis.
|
|
G.A.Gomez,
C.Morisseau,
B.D.Hammock,
D.W.Christianson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The X-ray crystal structure of human soluble epoxide hydrolase (sEH) has been
determined at 2.6 A resolution, revealing a domain-swapped quaternary structure
identical to that observed for the murine enzyme [Argiriadi, M. A., Morisseau,
C., Hammock, B. D., and Christianson, D. W. (1999) Proc. Natl. Acad. Sci. U.S.A.
96, 10637-10642]. As with the murine enzyme, the epoxide hydrolytic mechanism of
the human enzyme proceeds through an alkyl-enzyme intermediate with Asp-333 in
the C-terminal domain. The structure of the human sEH complex with
N-cyclohexyl-N'-(iodophenyl)urea (CIU) has been determined at 2.35 A resolution.
Tyr-381 and Tyr-465 donate hydrogen bonds to the alkylurea carbonyl group of
CIU, consistent with the proposed roles of these residues as proton donors in
the first step of catalysis. The N-terminal domain of mammalian sEH contains a
15 A deep cleft, but its biological function is unclear. Recent experiments
demonstrate that the N-terminal domain of human sEH catalyzes the
metal-dependent hydrolysis of phosphate esters [Cronin, A., Mowbray, S., Dürk,
H., Homburg, S., Fleming, I., Fisslthaler, B., Oesch, F., and Arand, M. (2003)
Proc. Natl. Acad. Sci. U.S.A. 100, 1552-1557; Newman, J. W., Morisseau, C.,
Harris, T. R., and Hammock, B. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100,
1558-1563]. The binding of Mg(2+)-HPO4(2-) to the N-terminal domain of human sEH
in its CIU complex reveals structural features relevant to those of the
enzyme-substrate complex in the phosphatase reaction.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Sheng,
C.Wei,
Z.Zhang,
S.Wang,
and
Q.Zhu
(2011).
Enantioselective hydrolysis of glycidyl methylphenyl ethers by Botryosphaeria dothidea ZJUZQ007: effect of substitution pattern on enantioselectivity.
|
| |
Appl Biochem Biotechnol,
164,
125-132.
|
 |
|
|
|
|
 |
M.Decker,
M.Arand,
and
A.Cronin
(2009).
Mammalian epoxide hydrolases in xenobiotic metabolism and signalling.
|
| |
Arch Toxicol,
83,
297-318.
|
 |
|
|
|
|
 |
M.W.Buczynski,
D.S.Dumlao,
and
E.A.Dennis
(2009).
Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology.
|
| |
J Lipid Res,
50,
1015-1038.
|
 |
|
|
|
|
 |
S.Lin,
G.P.Horsman,
Y.Chen,
W.Li,
and
B.Shen
(2009).
Characterization of the SgcF epoxide hydrolase supporting an (R)-vicinal diol intermediate for enediyne antitumor antibiotic C-1027 biosynthesis.
|
| |
J Am Chem Soc,
131,
16410-16417.
|
 |
|
|
|
|
 |
B.K.Biswal,
C.Morisseau,
G.Garen,
M.M.Cherney,
C.Garen,
C.Niu,
B.D.Hammock,
and
M.N.James
(2008).
The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor.
|
| |
J Mol Biol,
381,
897-912.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Melzer,
S.Junne,
R.Wohlgemuth,
D.C.Hempel,
and
P.Götz
(2008).
Production of epoxide hydrolases in batch fermentations of Botryosphaeria rhodina.
|
| |
J Ind Microbiol Biotechnol,
35,
485-493.
|
 |
|
|
|
|
 |
T.R.Harris,
P.A.Aronov,
and
B.D.Hammock
(2008).
Soluble epoxide hydrolase homologs in Strongylocentrotus purpuratus suggest a gene duplication event and subsequent divergence.
|
| |
DNA Cell Biol,
27,
467-477.
|
 |
|
|
|
|
 |
T.Y.Zakharian,
L.Di Costanzo,
and
D.W.Christianson
(2008).
Synthesis of (2S)-2-amino-7,8-epoxyoctanoic acid and structure of its metal-bridging complex with human arginase I.
|
| |
Org Biomol Chem,
6,
3240-3243.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Luria,
S.M.Weldon,
A.K.Kabcenell,
R.H.Ingraham,
D.Matera,
H.Jiang,
R.Gill,
C.Morisseau,
J.W.Newman,
and
B.D.Hammock
(2007).
Compensatory mechanism for homeostatic blood pressure regulation in Ephx2 gene-disrupted mice.
|
| |
J Biol Chem,
282,
2891-2898.
|
 |
|
|
|
|
 |
B.Inceoglu,
K.R.Schmelzer,
C.Morisseau,
S.L.Jinks,
and
B.D.Hammock
(2007).
Soluble epoxide hydrolase inhibition reveals novel biological functions of epoxyeicosatrienoic acids (EETs).
|
| |
Prostaglandins Other Lipid Mediat,
82,
42-49.
|
 |
|
|
|
|
 |
K.H.Kim
(2007).
Outliers in SAR and QSAR: is unusual binding mode a possible source of outliers?
|
| |
J Comput Aided Mol Des,
21,
63-86.
|
 |
|
|
|
|
 |
M.De Vivo,
B.Ensing,
M.Dal Peraro,
G.A.Gomez,
D.W.Christianson,
and
M.L.Klein
(2007).
Proton shuttles and phosphatase activity in soluble epoxide hydrolase.
|
| |
J Am Chem Soc,
129,
387-394.
|
 |
|
|
|
|
 |
N.Chiamvimonvat,
C.M.Ho,
H.J.Tsai,
and
B.D.Hammock
(2007).
The soluble epoxide hydrolase as a pharmaceutical target for hypertension.
|
| |
J Cardiovasc Pharmacol,
50,
225-237.
|
 |
|
|
|
|
 |
C.Morisseau,
J.W.Newman,
H.J.Tsai,
P.A.Baecker,
and
B.D.Hammock
(2006).
Peptidyl-urea based inhibitors of soluble epoxide hydrolases.
|
| |
Bioorg Med Chem Lett,
16,
5439-5444.
|
 |
|
|
|
|
 |
G.A.Gomez,
C.Morisseau,
B.D.Hammock,
and
D.W.Christianson
(2006).
Human soluble epoxide hydrolase: structural basis of inhibition by 4-(3-cyclohexylureido)-carboxylic acids.
|
| |
Protein Sci,
15,
58-64.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.H.Hopmann,
and
F.Himo
(2006).
Theoretical study of the full reaction mechanism of human soluble epoxide hydrolase.
|
| |
Chemistry,
12,
6898-6909.
|
 |
|
|
|
|
 |
P.D.Jones,
H.J.Tsai,
Z.N.Do,
C.Morisseau,
and
B.D.Hammock
(2006).
Synthesis and SAR of conformationally restricted inhibitors of soluble epoxide hydrolase.
|
| |
Bioorg Med Chem Lett,
16,
5212-5216.
|
 |
|
|
|
|
 |
S.L.Mowbray,
L.T.Elfström,
K.M.Ahlgren,
C.E.Andersson,
and
M.Widersten
(2006).
X-ray structure of potato epoxide hydrolase sheds light on substrate specificity in plant enzymes.
|
| |
Protein Sci,
15,
1628-1637.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.R.Harris,
C.Morisseau,
R.L.Walzem,
S.J.Ma,
and
B.D.Hammock
(2006).
The cloning and characterization of a soluble epoxide hydrolase in chicken.
|
| |
Poult Sci,
85,
278-287.
|
 |
|
|
|
|
 |
C.Morisseau,
and
B.D.Hammock
(2005).
Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles.
|
| |
Annu Rev Pharmacol Toxicol,
45,
311-333.
|
 |
|
|
|
|
 |
J.W.Newman,
C.Morisseau,
and
B.D.Hammock
(2005).
Epoxide hydrolases: their roles and interactions with lipid metabolism.
|
| |
Prog Lipid Res,
44,
1.
|
 |
|
|
|
|
 |
K.L.Tran,
P.A.Aronov,
H.Tanaka,
J.W.Newman,
B.D.Hammock,
and
C.Morisseau
(2005).
Lipid sulfates and sulfonates are allosteric competitive inhibitors of the N-terminal phosphatase activity of the mammalian soluble epoxide hydrolase.
|
| |
Biochemistry,
44,
12179-12187.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

