 |
PDBsum entry 1vch

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E:
E.C.2.4.2.7
- adenine phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Ribose activation
|
 |
 |
 |
 |
 |
Reaction:
|
 |
AMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + adenine
|
 |
 |
 |
 |
 |
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
=
|
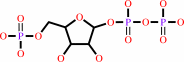 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
+
|
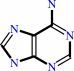 adenine
adenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
61:658-665
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a purine/pyrimidine phosphoribosyltransferase-related protein from Thermus thermophilus HB8.
|
|
P.H.Rehse,
T.H.Tahirov.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Adenine phosphoribosyltransferase (APRTase) is a widely distributed enzyme
involved in the salvage of adenine to form an adenine nucleotide. We
crystallized and determined the X-ray crystallographic structure of a
purine/pyrimidine phosphoribosyltransferase-related protein from the
thermophilic bacterium, Thermus thermophilus HB8. The crystal space group was C2
with unit cell dimensions of a = 167.42 A, b = 61.41 A, c = 102.39 A, beta =
94.0 degrees . Initial phases were determined to 2.6 A using the multiple
wavelength anomalous dispersion method and selenomethionine substituted protein
(Se-MAD), and refined using a 1.9 A "native" data set. The asymmetric
unit contains two pairs of identical dimers, each related by noncrystallographic
two-fold symmetry. The fifth monomer forms a similar dimer across a
crystallographic two-fold axis. These dimers appear to be the biological unit
with both monomers contributing to an unusual highly charged arginine-rich
bridge region separating the two active sites. Comparison with distantly related
APRTases reveal similarities and differences of the active site.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. (a) Stereo view of the biological unit. (b)
Electrostatic potential surface of TtAPRTase. (c) Electrostatic
potential surface of human APRTase.Both (a) and (b) are oriented
to display the arginine-rich bridging region of the TtAPRTase.
The AMP is modeled into the TtAPRTase.
|
 |
Figure 4.
Figure 4. (a) Active site including the arginine-rich region
created by the two-fold axis. The modeled AMP is in blue; the A
chain is in black, and the B chain is in red. (b) Selected
active site residues from the T. thermophilus (yellow) and human
(red) APRTases. AMP is in blue.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2005,
61,
658-665)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

