 |
PDBsum entry 1v79

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.4
- adenosine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
adenosine + H2O + H+ = inosine + NH4+
|
|
2.
|
2'-deoxyadenosine + H2O + H+ = 2'-deoxyinosine + NH4+
|
|
 |
 |
 |
 |
 |
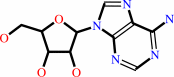 adenosine
adenosine
|
+
|
H2O
|
+
|
H(+)
|
=
|
inosine
Bound ligand (Het Group name = )
matches with 46.43% similarity
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
2'-deoxyadenosine
Bound ligand (Het Group name = )
matches with 42.86% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyinosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
47:2728-2731
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors.
|
|
T.Terasaka,
H.Okumura,
K.Tsuji,
T.Kato,
I.Nakanishi,
T.Kinoshita,
Y.Kato,
M.Kuno,
N.Seki,
Y.Naoe,
T.Inoue,
K.Tanaka,
K.Nakamura.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We disclose optimization efforts based on the novel non-nucleoside adenosine
deaminase (ADA) inhibitor, 4 (K(i) = 680 nM). Structure-based drug design
utilizing the crystal structure of the 4/ADA complex led to discovery of 5 (K(i)
= 11 nM, BA = 30% in rats). Furthermore, from metabolic considerations, we
discovered two inhibitors with improved oral bioavailability [6 (K(i) = 13 nM,
BA = 44%) and 7 (K(i) = 9.8 nM, BA = 42%)]. 6 demonstrated in vivo efficacy in
models of inflammation and lymphoma.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.T.Larson,
W.Deng,
B.E.Krumm,
A.Napuli,
N.Mueller,
W.C.Van Voorhis,
F.S.Buckner,
E.Fan,
A.Lauricella,
G.DeTitta,
J.Luft,
F.Zucker,
W.G.Hol,
C.L.Verlinde,
and
E.A.Merritt
(2008).
Structures of substrate- and inhibitor-bound adenosine deaminase from a human malaria parasite show a dramatic conformational change and shed light on drug selectivity.
|
| |
J Mol Biol,
381,
975-988.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.P.Williams,
L.F.Kuyper,
and
K.H.Pearce
(2005).
Recent applications of protein crystallography and structure-guided drug design.
|
| |
Curr Opin Chem Biol,
9,
371-380.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

