 |
PDBsum entry 2pgf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.5.4.31
- S-methyl-5'-thioadenosine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-methyl-5'-thioadenosine + H2O + H+ = S-methyl-5'-thioinosine + NH4+
|
 |
 |
 |
 |
 |
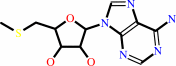 S-methyl-5'-thioadenosine
S-methyl-5'-thioadenosine
|
+
|
H2O
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
=
|
S-methyl-5'-thioinosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.4.4
- adenosine deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
adenosine + H2O + H+ = inosine + NH4+
|
|
2.
|
2'-deoxyadenosine + H2O + H+ = 2'-deoxyinosine + NH4+
|
|
 |
 |
 |
 |
 |
adenosine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
+
|
H(+)
|
=
|
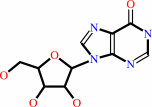 inosine
inosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
2'-deoxyadenosine
Bound ligand (Het Group name = )
matches with 94.74% similarity
|
+
|
H2O
|
+
|
H(+)
|
=
|
2'-deoxyinosine
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
381:975-988
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of substrate- and inhibitor-bound adenosine deaminase from a human malaria parasite show a dramatic conformational change and shed light on drug selectivity.
|
|
E.T.Larson,
W.Deng,
B.E.Krumm,
A.Napuli,
N.Mueller,
W.C.Van Voorhis,
F.S.Buckner,
E.Fan,
A.Lauricella,
G.DeTitta,
J.Luft,
F.Zucker,
W.G.Hol,
C.L.Verlinde,
E.A.Merritt.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Plasmodium and other apicomplexan parasites are deficient in purine
biosynthesis, relying instead on the salvage of purines from their host
environment. Therefore, interference with the purine salvage pathway is an
attractive therapeutic target. The plasmodial enzyme adenosine deaminase (ADA)
plays a central role in purine salvage and, unlike mammalian ADA homologs, has a
further secondary role in methylthiopurine recycling. For this reason,
plasmodial ADA accepts a wider range of substrates, as it is responsible for
deamination of both adenosine and 5'-methylthioadenosine. The latter substrate
is not accepted by mammalian ADA homologs. The structural basis for this natural
difference in specificity between plasmodial and mammalian ADA has not been well
understood. We now report crystal structures of Plasmodium vivax ADA in complex
with adenosine, guanosine, and the picomolar inhibitor 2'-deoxycoformycin. These
structures highlight a drastic conformational change in plasmodial ADA upon
substrate binding that has not been observed for mammalian ADA enzymes. Further,
these complexes illuminate the structural basis for the differential substrate
specificity and potential drug selectivity between mammalian and parasite
enzymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. The boot-shaped active-site cavity and putative
ammonium channel gate of the active conformation of plasmodial
ADA. (a) Side view of the cavity, looking into the side opposite
the catalytic zinc. The enclosed adenosine and DCF (yellow and
orange sticks, respectively) and water molecules (red spheres)
that occupy the cavity are displayed. The catalytic zinc
(magenta spheres) makes up one wall of the “heel” of the
boot. (b) The view has been rotated − 100° along the
y-axis and is now into the “toe” of the boot. Note that the
hydroxyl group of DCF that is equivalent to the leaving amine
group of adenosine is oriented toward the putative ammonium
channel. (c) The ammonia channel gate. Conformational changes in
α13 and in the side chain of Asp205 exist between the closed,
substrate-bound (d) and open, apo (e) forms of ADA. In the
closed form, the solvent-filled channel leading to the surface
from the active site is blocked by the side chain of Asp205.
When the enzyme is not bound to ligand, the Asp205 side chain
adopts an alternate conformation that allows the channel access
to the surrounding solvent, presumably facilitating the release
of the ammonia product.
|
 |
Figure 6.
Fig. 6. (a and b) Alternate sugar pucker of
substrate/inhibitor induced by the plasmodial ADA
Asp172:mammalian ADA Met155 sequence difference. Plasmodial ADA
is cyan and its bound DCF in orange, while mammalian ADA is
green and its bound DCF in pink. Plasmodial ADA Asp172
hydrogen-bonds with the ribose 3′-hydroxyl group, an
interaction that mammalian Met155 is incapable of making. This
causes the plasmodial ADA-bound inhibitor to adopt a C2′-endo
sugar pucker, while the mammalian ADA-bound inhibitor adopts a
C4′-exo pucker. The result is that the 5′-carbon of the two
riboses are oriented significantly differently with respect to
the ribose ring, although the 5′-hydroxyl groups occupy nearly
the same location and are less than 0.4 Å apart. The
different orientations of the 5′-carbon, however, has a great
affect on the space that additions at this position may occupy,
while maintaining a biologically relevant glycosidic linkage
with the purine ring. (c) Stereo view of 5′-PhS-DCF (purple
sticks) docked into the active-site cavity of plasmodial ADA and
superimposed on the crystallographically observed DCF (orange
sticks). The plasmodial ADA crystal structure is cyan, while the
protein following docking is green. The most significant change
in the structure of plasmodial ADA in order to accommodate the
5′-thiophenyl addition is an alternate rotamer adopted by
Phe132, which both enlarges the cavity and stabilizes the 5′
addition.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2008,
381,
975-988)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.V.Zavialov,
X.Yu,
D.Spillmann,
G.Lauvau,
and
A.V.Zavialov
(2010).
Structural basis for the growth factor activity of human adenosine deaminase ADA2.
|
| |
J Biol Chem,
285,
12367-12377.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.C.Ho,
M.B.Cassera,
D.C.Madrid,
L.M.Ting,
P.C.Tyler,
K.Kim,
S.C.Almo,
and
V.L.Schramm
(2009).
Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases.
|
| |
Biochemistry,
48,
9618-9626.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.N.Wells,
P.L.Alonso,
and
W.E.Gutteridge
(2009).
New medicines to improve control and contribute to the eradication of malaria.
|
| |
Nat Rev Drug Discov,
8,
879-891.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

