 |
PDBsum entry 1v4k

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.11
- Transferred entry: 2.5.1.84 and 2.5.1.85.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Terpenoid biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
All-trans-octaprenyl diphosphate + isopentenyl diphosphate = diphosphate + all-trans-nonaprenyl diphosphate
|
 |
 |
 |
 |
 |
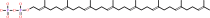 All-trans-octaprenyl diphosphate
All-trans-octaprenyl diphosphate
|
+
|
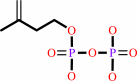 isopentenyl diphosphate
isopentenyl diphosphate
|
=
|
 diphosphate
diphosphate
|
+
|
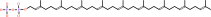 all-trans-nonaprenyl diphosphate
all-trans-nonaprenyl diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:4903-4912
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of octaprenyl pyrophosphate synthase from hyperthermophilic Thermotoga maritima and mechanism of product chain length determination.
|
|
R.T.Guo,
C.J.Kuo,
C.C.Chou,
T.P.Ko,
H.L.Shr,
P.H.Liang,
A.H.Wang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Octaprenyl pyrophosphate synthase (OPPs) catalyzes consecutive condensation
reactions of farnesyl pyrophosphate (FPP) with isopentenyl pyrophosphate (IPP)
to generate C40 octaprenyl pyrophosphate (OPP), which constitutes the side chain
of bacterial ubiquinone or menaquinone. In this study, the first structure of
long chain C40-OPPs from Thermotoga maritima has been determined to 2.28-A
resolution. OPPs is composed entirely of alpha-helices joined by connecting
loops and is arranged with nine core helices around a large central cavity. An
elongated hydrophobic tunnel between D and F alpha-helices contains two DDXXD
motifs on the top for substrate binding and is occupied at the bottom with two
large residues Phe-52 and Phe-132. The products of the mutant F132A OPPs are
predominantly C50, longer than the C40 synthesized by the wild-type and F52A
mutant OPPs, suggesting that Phe-132 is the key residue for determining the
product chain length. Ala-76 and Ser-77 located close to the FPP binding site
and Val-73 positioned further down the tunnel were individually mutated to
larger amino acids. A76Y and S77F mainly produce C20 indicating that the mutated
large residues in the vicinity of the FPP site limit the substrate chain
elongation. Ala-76 is the fifth amino acid upstream from the first DDXXD motif
on helix D of OPPs, and its corresponding amino acid in FPPs is Tyr. In
contrast, V73Y mutation led to additional accumulation of C30 intermediate. The
new structure of the trans-type OPPs, together with the recently determined
cis-type UPPs, significantly extends our understanding on the biosynthesis of
long chain polyprenyl molecules.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIG. 3. OPPs active site structure and reaction mechanism.
In A, the surface of active site is color coded from red to blue
according to charge potential from -15 to 15 k[B]T. This figure
was generated using GRASP (30). In B, two sulfate ions in the
active site of totally six sulfates of OPPs F132A mutant are
shown (these sulfate ions are more obvious than other data
sets). Along with two sulfate ions, amino acids Lys-41, Arg-44,
His-74, Asp-81, Asp-82, Asp-85, Arg-90, Arg-91, Asp-204,
Asp-205, and Asp-208 are shown in ball-and-stick model. S1
containing the first DDXXD motif is responsible for binding with
FPP, and S2 located downstream the FPP binding site functions to
stabilize the PP[i] leaving group. This is illustrated in C that
Arg-90 and Arg-91 are important in FPP binding, and AArg-44,
Lys-41, and His-74 surround another sulfate ion to grasp the
leaving group of FPP while reaction occurs.
|
 |
Figure 8.
FIG. 8. Proposed mechanism for chain length determination
catalyzed by OPPs. The first DDXXD motif attached to helix D
represents the FPP binding site. Ser-77 and Ala-76 are located
in immediate proximity of FPP, and V73Y is further down. The
substitution of Ser-77 and Ala-76 with larger residues led to
the formation of C[20], a single condensation between the bound
FPP and IPP. V73Y mutation results in temporary accumulation of
C[30]. Phe-132 located on the bottom of helix D blocks further
chain elongation of OPP and determines the ultimate product
chain length.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
4903-4912)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.H.Chang,
F.L.Hsieh,
T.P.Ko,
K.H.Teng,
P.H.Liang,
and
A.H.Wang
(2010).
Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation.
|
| |
Plant Cell,
22,
454-467.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Kawamukai
(2009).
Biosynthesis and bioproduction of coenzyme Q10 by yeasts and other organisms.
|
| |
Biotechnol Appl Biochem,
53,
217-226.
|
 |
|
|
|
|
 |
J.D.Artz,
J.E.Dunford,
M.J.Arrowood,
A.Dong,
M.Chruszcz,
K.L.Kavanagh,
W.Minor,
R.G.Russell,
F.H.Ebetino,
U.Oppermann,
and
R.Hui
(2008).
Targeting a uniquely nonspecific prenyl synthase with bisphosphonates to combat cryptosporidiosis.
|
| |
Chem Biol,
15,
1296-1306.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Noike,
T.Katagiri,
T.Nakayama,
T.Koyama,
T.Nishino,
and
H.Hemmi
(2008).
The product chain length determination mechanism of type II geranylgeranyl diphosphate synthase requires subunit interaction.
|
| |
FEBS J,
275,
3921-3933.
|
 |
|
|
|
|
 |
M.Zhang,
J.Luo,
Y.Ogiyama,
R.Saiki,
and
M.Kawamukai
(2008).
Heteromer formation of a long-chain prenyl diphosphate synthase from fission yeast Dps1 and budding yeast Coq1.
|
| |
FEBS J,
275,
3653-3668.
|
 |
|
|
|
|
 |
R.Saiki,
A.L.Lunceford,
Y.Shi,
B.Marbois,
R.King,
J.Pachuski,
M.Kawamukai,
D.L.Gasser,
and
C.F.Clarke
(2008).
Coenzyme Q10 supplementation rescues renal disease in Pdss2kd/kd mice with mutations in prenyl diphosphate synthase subunit 2.
|
| |
Am J Physiol Renal Physiol,
295,
F1535-F1544.
|
 |
|
|
|
|
 |
D.Umeno,
A.V.Tobias,
and
F.H.Arnold
(2005).
Diversifying carotenoid biosynthetic pathways by directed evolution.
|
| |
Microbiol Mol Biol Rev,
69,
51-78.
|
 |
|
|
|
|
 |
H.Y.Sun,
T.P.Ko,
C.J.Kuo,
R.T.Guo,
C.C.Chou,
P.H.Liang,
and
A.H.Wang
(2005).
Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures.
|
| |
J Bacteriol,
187,
8137-8148.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Saiki,
A.Nagata,
T.Kainou,
H.Matsuda,
and
M.Kawamukai
(2005).
Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans.
|
| |
FEBS J,
272,
5606-5622.
|
 |
|
|
|
|
 |
M.A.Pysz,
S.B.Conners,
C.I.Montero,
K.R.Shockley,
M.R.Johnson,
D.E.Ward,
and
R.M.Kelly
(2004).
Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima.
|
| |
Appl Environ Microbiol,
70,
6098-6112.
|
 |
|
|
|
|
 |
S.Y.Chang,
T.P.Ko,
A.P.Chen,
A.H.Wang,
and
P.H.Liang
(2004).
Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies.
|
| |
Protein Sci,
13,
971-978.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

