 |
PDBsum entry 1udw

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of human uridine-cytidine kinase 2 complexed with a feedback-inhibitor, ctp
|
|
Structure:
|
 |
Uridine-cytidine kinase 2. Chain: a, b. Fragment: residues 1-250. Synonym: uridine-cytidine kinase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693.
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.210
|
R-free:
|
0.251
|
|
|
Authors:
|
 |
N.N.Suzuki,K.Koizumi,M.Fukushima,A.Matsuda,F.Inagaki
|
Key ref:

|
 |
N.N.Suzuki
et al.
(2004).
Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase.
Structure,
12,
751-764.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
07-May-03
|
Release date:
|
04-May-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9BZX2
(UCK2_HUMAN) -
Uridine-cytidine kinase 2 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
261 a.a.
204 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.48
- uridine/cytidine kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
uridine + ATP = UMP + ADP + H+
|
|
2.
|
cytidine + ATP = CMP + ADP + H+
|
|
 |
 |
 |
 |
 |
uridine
Bound ligand (Het Group name = )
matches with 87.50% similarity
|
+
|
 ATP
ATP
|
=
|
 UMP
UMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
cytidine
Bound ligand (Het Group name = )
matches with 87.50% similarity
|
+
|
 ATP
ATP
|
=
|
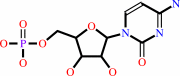 CMP
CMP
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
12:751-764
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase.
|
|
N.N.Suzuki,
K.Koizumi,
M.Fukushima,
A.Matsuda,
F.Inagaki.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Uridine-cytidine kinase (UCK) catalyzes the phosphorylation of uridine and
cytidine and activates pharmacological ribonucleoside analogs. Here we present
the crystal structures of human UCK alone and in complexes with a substrate,
cytidine, a feedback inhibitor, CTP or UTP, and with phosphorylation products,
CMP and ADP, respectively. Free UCK takes an alpha/beta mononucleotide binding
fold and exists as a homotetramer with 222 symmetry. Upon inhibitor binding, one
loop region was loosened, causing the UCK tetramer to be distorted. Upon
cytidine binding, a large induced fit was observed at the uridine/cytidine
binding site, which endows UCK with a strict specificity for pyrimidine
ribonucleosides. The first UCK structure provided the structural basis for the
specificity, catalysis, and regulation of human uridine-cytidine kinase, which
give clues for the design of novel antitumor and antiviral ribonucleoside
analogs that inhibit RNA synthesis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 3.
Figure 3. Final Annealed F[o] - F[c] Omit Electron Density
Map for Ligands Bound to UCK(A) CMP, ADP, and the magnesium ion
observed in the CMP-ADP-bound UCK crystal. The map is contoured
at 3.5 s and the resolution is 1.8 Å.(B) CTP observed in the
CTP-bound UCK crystal. The map is contoured at 4.0 s and the
resolution is 2.6 Å.(C) UTP observed in UTP-bound UCK. The map
is contoured at 4.0 s and the resolution is 2.6 Å.(D) Cyd and
citrate observed in Cyd-bound UCK. The map is contoured at 3.5 s
and the resolution is 2.6 Å. This figure was prepared using
program O (Jones et al., 1991).
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2004,
12,
751-764)
copyright 2004.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.J.Smith,
Y.Li,
and
K.N.Houk
(2009).
Quantum mechanics/molecular mechanics investigation of the mechanism of phosphate transfer in human uridine-cytidine kinase 2.
|
| |
Org Biomol Chem,
7,
2716-2724.
|
 |
|
|
|
|
 |
A.Lossani,
A.Torti,
S.Gatti,
A.Bruno,
R.Maserati,
G.E.Wright,
and
F.Focher
(2009).
Thymidine kinase and uridine-cytidine kinase from Entamoeba histolytica: cloning, characterization, and search for specific inhibitors.
|
| |
Parasitology,
136,
595-602.
|
 |
|
|
|
|
 |
P.J.Kamphuis,
and
R.J.Wurtman
(2009).
Nutrition and Alzheimer's disease: pre-clinical concepts.
|
| |
Eur J Neurol,
16,
12-18.
|
 |
|
|
|
|
 |
Z.B.Redzic,
S.A.Malatiali,
J.D.Craik,
M.L.Rakic,
and
A.J.Isakovic
(2009).
Blood-brain barrier efflux transport of pyrimidine nucleosides and nucleobases in the rat.
|
| |
Neurochem Res,
34,
566-573.
|
 |
|
|
|
|
 |
S.Holguin,
J.Martinez,
C.Chow,
and
R.Wurtman
(2008).
Dietary uridine enhances the improvement in learning and memory produced by administering DHA to gerbils.
|
| |
FASEB J,
22,
3938-3946.
|
 |
|
|
|
|
 |
S.Holguin,
Y.Huang,
J.Liu,
and
R.Wurtman
(2008).
Chronic administration of DHA and UMP improves the impaired memory of environmentally impoverished rats.
|
| |
Behav Brain Res,
191,
11-16.
|
 |
|
|
|
|
 |
V.Gueguen-Chaignon,
V.Chaptal,
L.Larivière,
N.Costa,
P.Lopes,
S.Morera,
and
S.Nessler
(2008).
Crystal structure and functional analysis identify the P-loop containing protein YFH7 of Saccharomyces cerevisiae as an ATP-dependent kinase.
|
| |
Proteins,
71,
804-812.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Murakami,
H.Bao,
M.Ramesh,
T.R.McBrayer,
T.Whitaker,
H.M.Micolochick Steuer,
R.F.Schinazi,
L.J.Stuyver,
A.Obikhod,
M.J.Otto,
and
P.A.Furman
(2007).
Mechanism of activation of beta-D-2'-deoxy-2'-fluoro-2'-c-methylcytidine and inhibition of hepatitis C virus NS5B RNA polymerase.
|
| |
Antimicrob Agents Chemother,
51,
503-509.
|
 |
|
|
|
|
 |
K.Walldén,
P.Stenmark,
T.Nyman,
S.Flodin,
S.Gräslund,
P.Loppnau,
V.Bianchi,
and
P.Nordlund
(2007).
Crystal structure of human cytosolic 5'-nucleotidase II: insights into allosteric regulation and substrate recognition.
|
| |
J Biol Chem,
282,
17828-17836.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Wang,
M.A.Albrecht,
and
R.J.Wurtman
(2007).
Dietary supplementation with uridine-5'-monophosphate (UMP), a membrane phosphatide precursor, increases acetylcholine level and release in striatum of aged rat.
|
| |
Brain Res,
1133,
42-48.
|
 |
|
|
|
|
 |
W.Tempel,
W.M.Rabeh,
K.L.Bogan,
P.Belenky,
M.Wojcik,
H.F.Seidle,
L.Nedyalkova,
T.Yang,
A.A.Sauve,
H.W.Park,
and
C.Brenner
(2007).
Nicotinamide riboside kinase structures reveal new pathways to NAD+.
|
| |
PLoS Biol,
5,
e263.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Ichikawa,
N.Minakawa,
S.Shuto,
M.Tanaka,
T.Sasaki,
and
A.Matsuda
(2006).
Synthesis of 3'-beta-carbamoylmethylcytidine (CAMC) and its derivatives as potential antitumor agents.
|
| |
Org Biomol Chem,
4,
1284-1296.
|
 |
|
|
|
|
 |
T.C.Appleby,
G.Larson,
I.W.Cheney,
H.Walker,
J.Z.Wu,
W.Zhong,
Z.Hong,
and
N.Yao
(2005).
Structure of human uridine-cytidine kinase 2 determined by SIRAS using a rotating-anode X-ray generator and a single samarium derivative.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
278-284.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

