 |
PDBsum entry 1trk

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase(ketone residues)
|
PDB id
|
|

|

|
1trk
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.2.1.1
- transketolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = aldehydo-D- ribose 5-phosphate + D-xylulose 5-phosphate
|
 |
 |
 |
 |
 |
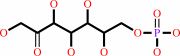 D-sedoheptulose 7-phosphate
D-sedoheptulose 7-phosphate
|
+
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
=
|
aldehydo-D- ribose 5-phosphate
|
+
|
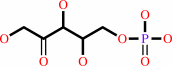 D-xylulose 5-phosphate
D-xylulose 5-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
TPP)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
238:387-404
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Refined structure of transketolase from Saccharomyces cerevisiae at 2.0 A resolution.
|
|
M.Nikkola,
Y.Lindqvist,
G.Schneider.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of transketolase from Saccharomyces cerevisiae has been
refined to a crystallographic residual of 15.7% at 2.0 A resolution using the
program package X-PLOR. The refined model of the transketolase homodimer,
corresponding to 1356 amino acid residues in the asymmetric unit, consists of
10,396 protein atoms, 1040 solvent molecules, 52 thiamine diphosphate atoms and
two calcium ions. All amino acid residues except for the two N-terminal residues
of the two subunits are defined in the electron density maps and refined. The
estimated root-mean-square (r.m.s.) error of the model is less than 0.2 A as
deduced from Luzzati plots. The r.m.s. deviation from ideality is 0.017 A for
bond distances and 3.1 degrees for bond angles. The main-chain torsion angles of
non-glycine residues lie within the allowed regions of the Ramachandran plots.
The model shows a very good fit to the electron density maps. The average
B-factor for all protein atoms in the first subunit is 19 A2, and 15A2 in the
second. The average B-factor for solvent atoms is 32A2. The two subunits of
transketolase were refined independently and have nearly identical structures
with an r.m.s. deviation of 0.24 A for C alpha atoms 3 to 680, and slightly less
when aligning the individual domains. A few exceptions from the 2-fold symmetry
are found, mostly in the surface residues. The thiamine diphosphate cofactors
have identical conformations. The cofactor is shielded from solvent except for
the C-2 atom of the thiazolium ring. A calcium ion is bound to the diphosphate
group of thiamine and protein ligands. The metal binding site and the
interactions of thiamine diphosphate with protein residues are described. A
network of hydrogen bonds consisting of glutamic acid residues and internal
water molecules connects the two thiamine diphosphate molecules. Its structure
and possible functional implications are discussed.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.A.Kochetov,
and
I.A.Sevostyanova
(2010).
Functional nonequivalence of transketolase active centers.
|
| |
IUBMB Life,
62,
797-802.
|
 |
|
|
|
|
 |
L.E.Meshalkina,
O.N.Solovjeva,
Y.A.Khodak,
V.L.Drutsa,
and
G.A.Kochetov
(2010).
Isolation and properties of human transketolase.
|
| |
Biochemistry (Mosc),
75,
873-880.
|
 |
|
|
|
|
 |
X.Y.Pei,
K.M.Erixon,
B.F.Luisi,
and
F.J.Leeper
(2010).
Structural insights into the prereaction state of pyruvate decarboxylase from Zymomonas mobilis .
|
| |
Biochemistry,
49,
1727-1736.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.T.Jurgenson,
T.P.Begley,
and
S.E.Ealick
(2009).
The structural and biochemical foundations of thiamin biosynthesis.
|
| |
Annu Rev Biochem,
78,
569-603.
|
 |
|
|
|
|
 |
J.Zhao,
and
C.J.Zhong
(2009).
A review on research progress of transketolase.
|
| |
Neurosci Bull,
25,
94-99.
|
 |
|
|
|
|
 |
K.H.Sippel,
A.H.Robbins,
R.Reutzel,
S.K.Boehlein,
K.Namiki,
S.Goodison,
M.Agbandje-McKenna,
C.J.Rosser,
and
R.McKenna
(2009).
Structural insights into the extracytoplasmic thiamine-binding lipoprotein p37 of Mycoplasma hyorhinis.
|
| |
J Bacteriol,
191,
2585-2592.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.A.Esakova,
L.E.Meshalkina,
G.A.Kochetov,
and
R.Golbik
(2009).
Halogenated pyruvate derivatives as substrates of transketolase from Saccharomyces cerevisiae.
|
| |
Biochemistry (Mosc),
74,
1234-1238.
|
 |
|
|
|
|
 |
I.A.Sevostyanova,
V.A.Yurshev,
O.N.Solovjeva,
S.V.Zabrodskaya,
and
G.A.Kochetov
(2008).
Effect of bivalent cations on the interaction of transketolase with its donor substrate.
|
| |
Proteins,
71,
541-545.
|
 |
|
|
|
|
 |
J.P.Aucamp,
R.J.Martinez-Torres,
E.G.Hibbert,
and
P.A.Dalby
(2008).
A microplate-based evaluation of complex denaturation pathways: structural stability of Escherichia coli transketolase.
|
| |
Biotechnol Bioeng,
99,
1303-1310.
|
 |
|
|
|
|
 |
S.J.Costelloe,
J.M.Ward,
and
P.A.Dalby
(2008).
Evolutionary Analysis of the TPP-Dependent Enzyme Family.
|
| |
J Mol Evol,
66,
36-49.
|
 |
|
|
|
|
 |
R.V.Ospanov,
G.A.Kochetov,
and
B.I.Kurganov
(2007).
Influence of donor substrate on kinetic parameters of thiamine diphosphate binding to transketolase.
|
| |
Biochemistry (Mosc),
72,
84-92.
|
 |
|
|
|
|
 |
S.Xiang,
G.Usunow,
G.Lange,
M.Busch,
and
L.Tong
(2007).
Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis.
|
| |
J Biol Chem,
282,
2676-2682.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Malandrinos,
M.Louloudi,
and
N.Hadjiliadis
(2006).
Thiamine models and perspectives on the mechanism of action of thiamine-dependent enzymes.
|
| |
Chem Soc Rev,
35,
684-692.
|
 |
|
|
|
|
 |
O.A.Esakova,
L.E.Meshalkina,
and
G.A.Kochetov
(2005).
Effects of transketolase cofactors on its conformation and stability.
|
| |
Life Sci,
78,
8.
|
 |
|
|
|
|
 |
R.Golbik,
L.E.Meshalkina,
T.Sandalova,
K.Tittmann,
E.Fiedler,
H.Neef,
S.König,
R.Kluger,
G.A.Kochetov,
G.Schneider,
and
G.Hübner
(2005).
Effect of coenzyme modification on the structural and catalytic properties of wild-type transketolase and of the variant E418A from Saccharomyces cerevisiae.
|
| |
FEBS J,
272,
1326-1342.
|
 |
|
|
|
|
 |
O.A.Esakova,
L.E.Meshalkina,
R.Golbik,
G.Hübner,
and
G.A.Kochetov
(2004).
Donor substrate regulation of transketolase.
|
| |
Eur J Biochem,
271,
4189-4194.
|
 |
|
|
|
|
 |
R.A.Frank,
C.M.Titman,
J.V.Pratap,
B.F.Luisi,
and
R.N.Perham
(2004).
A molecular switch and proton wire synchronize the active sites in thiamine enzymes.
|
| |
Science,
306,
872-876.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Fiedler,
S.Thorell,
T.Sandalova,
R.Golbik,
S.König,
and
G.Schneider
(2002).
Snapshot of a key intermediate in enzymatic thiamin catalysis: crystal structure of the alpha-carbanion of (alpha,beta-dihydroxyethyl)-thiamin diphosphate in the active site of transketolase from Saccharomyces cerevisiae.
|
| |
Proc Natl Acad Sci U S A,
99,
591-595.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.S.Pang,
L.W.Guddat,
and
R.G.Duggleby
(2001).
Crystallization of the catalytic subunit of Saccharomyces cerevisiae acetohydroxyacid synthase.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1321-1323.
|
 |
|
|
|
|
 |
A.AEvarsson,
J.L.Chuang,
R.M.Wynn,
S.Turley,
D.T.Chuang,
and
W.G.Hol
(2000).
Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease.
|
| |
Structure,
8,
277-291.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.I.Svergun,
M.V.Petoukhov,
M.H.Koch,
and
S.König
(2000).
Crystal versus solution structures of thiamine diphosphate-dependent enzymes.
|
| |
J Biol Chem,
275,
297-302.
|
 |
|
|
|
|
 |
N.J.Turner
(2000).
Applications of transketolases in organic synthesis.
|
| |
Curr Opin Biotechnol,
11,
527-531.
|
 |
|
|
|
|
 |
H.J.Chiu,
J.J.Reddick,
T.P.Begley,
and
S.E.Ealick
(1999).
Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 A resolution.
|
| |
Biochemistry,
38,
6460-6470.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.H.Charon,
A.Volbeda,
E.Chabriere,
L.Pieulle,
and
J.C.Fontecilla-Camps
(1999).
Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase.
|
| |
Curr Opin Struct Biol,
9,
663-669.
|
 |
|
|
|
|
 |
G.Schenk,
R.G.Duggleby,
and
P.F.Nixon
(1998).
Heterologous expression of human transketolase.
|
| |
Int J Biochem Cell Biol,
30,
369-378.
|
 |
|
|
|
|
 |
G.Schenk,
R.G.Duggleby,
and
P.F.Nixon
(1998).
Properties and functions of the thiamin diphosphate dependent enzyme transketolase.
|
| |
Int J Biochem Cell Biol,
30,
1297-1318.
|
 |
|
|
|
|
 |
G.Schneider,
and
Y.Lindqvist
(1998).
Crystallography and mutagenesis of transketolase: mechanistic implications for enzymatic thiamin catalysis.
|
| |
Biochim Biophys Acta,
1385,
387-398.
|
 |
|
|
|
|
 |
M.S.Hasson,
A.Muscate,
M.J.McLeish,
L.S.Polovnikova,
J.A.Gerlt,
G.L.Kenyon,
G.A.Petsko,
and
D.Ringe
(1998).
The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
|
| |
Biochemistry,
37,
9918-9930.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.M.Wynn,
J.R.Davie,
J.L.Chuang,
C.D.Cote,
and
D.T.Chuang
(1998).
Impaired assembly of E1 decarboxylase of the branched-chain alpha-ketoacid dehydrogenase complex in type IA maple syrup urine disease.
|
| |
J Biol Chem,
273,
13110-13118.
|
 |
|
|
|
|
 |
B.B.McConnell,
B.Burkholder,
and
D.J.Danner
(1997).
Two new mutations in the human E1 beta subunit of branched chain alpha-ketoacid dehydrogenase associated with maple syrup urine disease.
|
| |
Biochim Biophys Acta,
1361,
263-271.
|
 |
|
|
|
|
 |
G.Schenk,
F.J.Leeper,
R.England,
P.F.Nixon,
and
R.G.Duggleby
(1997).
The role of His113 and His114 in pyruvate decarboxylase from Zymomonas mobilis.
|
| |
Eur J Biochem,
248,
63-71.
|
 |
|
|
|
|
 |
L.Meshalkina,
U.Nilsson,
C.Wikner,
T.Kostikowa,
and
G.Schneider
(1997).
Examination of the thiamin diphosphate binding site in yeast transketolase by site-directed mutagenesis.
|
| |
Eur J Biochem,
244,
646-652.
|
 |
|
|
|
|
 |
R.A.Harris,
J.W.Hawes,
K.M.Popov,
Y.Zhao,
Y.Shimomura,
J.Sato,
J.Jaskiewicz,
and
T.D.Hurley
(1997).
Studies on the regulation of the mitochondrial alpha-ketoacid dehydrogenase complexes and their kinases.
|
| |
Adv Enzyme Regul,
37,
271-293.
|
 |
|
|
|
|
 |
T.Schiött,
C.von Wachenfeldt,
and
L.Hederstedt
(1997).
Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis.
|
| |
J Bacteriol,
179,
1962-1973.
|
 |
|
|
|
|
 |
U.Nilsson,
L.Meshalkina,
Y.Lindqvist,
and
G.Schneider
(1997).
Examination of substrate binding in thiamin diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis.
|
| |
J Biol Chem,
272,
1864-1869.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Flechner,
U.Dressen,
P.Westhoff,
K.Henze,
C.Schnarrenberger,
and
W.Martin
(1996).
Molecular characterization of transketolase (EC 2.2.1.1) active in the Calvin cycle of spinach chloroplasts.
|
| |
Plant Mol Biol,
32,
475-484.
|
 |
|
|
|
|
 |
C.French,
and
J.M.Ward
(1996).
Production and modification of E. coli transketolase for large-scale biocatalysis.
|
| |
Ann N Y Acad Sci,
799,
11-18.
|
 |
|
|
|
|
 |
C.K.Singleton,
J.J.Wang,
L.Shan,
and
P.R.Martin
(1996).
Conserved residues are functionally distinct within transketolases of different species.
|
| |
Biochemistry,
35,
15865-15869.
|
 |
|
|
|
|
 |
C.Wikner,
L.Meshalkina,
U.Nilsson,
S.Bäckström,
Y.Lindqvist,
and
G.Schneider
(1995).
His103 in yeast transketolase is required for substrate recognition and catalysis.
|
| |
Eur J Biochem,
233,
750-755.
|
 |
|
|
|
|
 |
M.S.Hasson,
A.Muscate,
G.T.Henehan,
P.F.Guidinger,
G.A.Petsko,
D.Ringe,
and
G.L.Kenyon
(1995).
Purification and crystallization of benzoylformate decarboxylase.
|
| |
Protein Sci,
4,
955-959.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

