 |
PDBsum entry 1so0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.3.3
- aldose 1-epimerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-glucose = beta-D-glucose
|
 |
 |
 |
 |
 |
alpha-D-glucose
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
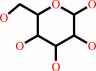 beta-D-glucose
beta-D-glucose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:23431-23437
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Molecular structure of human galactose mutarotase.
|
|
J.B.Thoden,
D.J.Timson,
R.J.Reece,
H.M.Holden.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Galactose mutarotase catalyzes the conversion of beta-d-galactose to
alpha-d-galactose during normal galactose metabolism. The enzyme has been
isolated from bacteria, plants, and animals and is present in the cytoplasm of
most cells. Here we report the x-ray crystallographic analysis of human
galactose mutarotase both in the apoform and complexed with its substrate,
beta-d-galactose. The polypeptide chain folds into an intricate array of 29
beta-strands, 25 classical reverse turns, and 2 small alpha-helices. There are
two cis-peptide bonds at Arg-78 and Pro-103. The sugar ligand sits in a shallow
cleft and is surrounded by Asn-81, Arg-82, His-107, His-176, Asp-243, Gln-279,
and Glu-307. Both the side chains of Glu-307 and His-176 are in the proper
location to act as a catalytic base and a catalytic acid, respectively. These
residues are absolutely conserved among galactose mutarotases. To date, x-ray
models for three mutarotases have now been reported, namely that described here
and those from Lactococcus lactis and Caenorhabditis elegans. The molecular
architectures of these enzymes differ primarily in the loop regions connecting
the first two beta-strands. In the human protein, there are six extra residues
in the loop compared with the bacterial protein for an approximate longer length
of 9 A. In the C. elegans protein, the first 17 residues are missing, thereby
reducing the total number of beta-strands by one.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
FIG. 2. Structure of human galactose mutarotase. A ribbon
representation of the three-dimensional architecture exhibited
by the apoenzyme is shown in a. For clarity, only the major
strands of the  -sheet are displayed.
Asterisks indicate the positions of cis-Arg-78 and cis-Pro-103.
The location of the active site is depicted in b as a
space-filling representation. Note that the 3-, 4-, and
6-hydroxyl groups of galactose, from left to right and indicated
by the red spheres, are solvent-exposed. The view is
approximately the same as displayed in a. -sheet are displayed.
Asterisks indicate the positions of cis-Arg-78 and cis-Pro-103.
The location of the active site is depicted in b as a
space-filling representation. Note that the 3-, 4-, and
6-hydroxyl groups of galactose, from left to right and indicated
by the red spheres, are solvent-exposed. The view is
approximately the same as displayed in a.
|
 |
Figure 3.
FIG. 3. The active site for human galactose mutarotase. A
close-up stereo view of those amino acid residues involved in
sugar binding is displayed in a. The color coding for the ribbon
representation is the same as in Fig. 2. The sugar is
highlighted in grayish bonds. Possible hydrogen bonding
interactions between the protein and the ligand are shown
schematically in b. The dotted lines indicate distances equal to
or below 3.2 Å.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
23431-23437)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Bakolitsa,
A.Kumar,
D.McMullan,
S.S.Krishna,
M.D.Miller,
D.Carlton,
R.Najmanovich,
P.Abdubek,
T.Astakhova,
H.J.Chiu,
T.Clayton,
M.C.Deller,
L.Duan,
Y.Elias,
J.Feuerhelm,
J.C.Grant,
S.K.Grzechnik,
G.W.Han,
L.Jaroszewski,
K.K.Jin,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
D.Marciano,
A.T.Morse,
E.Nigoghossian,
L.Okach,
S.Oommachen,
J.Paulsen,
R.Reyes,
C.L.Rife,
C.V.Trout,
H.van den Bedem,
D.Weekes,
A.White,
Q.Xu,
K.O.Hodgson,
J.Wooley,
M.A.Elsliger,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
and
I.A.Wilson
(2010).
The structure of the first representative of Pfam family PF06475 reveals a new fold with possible involvement in glycolipid metabolism.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1211-1217.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Lai,
L.J.Elsas,
and
K.J.Wierenga
(2009).
Galactose toxicity in animals.
|
| |
IUBMB Life,
61,
1063-1074.
|
 |
|
|
|
|
 |
A.Scott,
and
D.J.Timson
(2007).
Characterization of the Saccharomyces cerevisiae galactose mutarotase/UDP-galactose 4-epimerase protein, Gal10p.
|
| |
FEMS Yeast Res,
7,
366-371.
|
 |
|
|
|
|
 |
T.Pai,
Q.Chen,
Y.Zhang,
R.Zolfaghari,
and
A.C.Ross
(2007).
Galactomutarotase and other galactose-related genes are rapidly induced by retinoic acid in human myeloid cells.
|
| |
Biochemistry,
46,
15198-15207.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

