 |
PDBsum entry 1s3h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.3.1
- methylmalonyl-CoA carboxytransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-methylmalonyl-CoA + pyruvate = propanoyl-CoA + oxaloacetate
|
 |
 |
 |
 |
 |
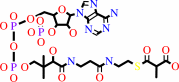 (S)-methylmalonyl-CoA
(S)-methylmalonyl-CoA
|
+
|
 pyruvate
pyruvate
|
=
|
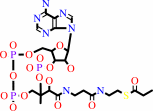 propanoyl-CoA
propanoyl-CoA
|
+
|
 oxaloacetate
oxaloacetate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Biotin; Cobalt cation; Zn(2+)
|
 |
 |
 |
 |
 |
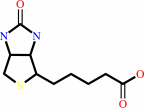 Biotin
Biotin
|
Cobalt cation
|
Zn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
23:3621-3631
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
|
|
P.R.Hall,
R.Zheng,
L.Antony,
M.Pusztai-Carey,
P.R.Carey,
V.C.Yee.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Transcarboxylase is a 1.2 million Dalton (Da) multienzyme complex from
Propionibacterium shermanii that couples two carboxylation reactions,
transferring CO(2)(-) from methylmalonyl-CoA to pyruvate to yield propionyl-CoA
and oxaloacetate. Crystal structures of the 5S metalloenzyme subunit, which
catalyzes the second carboxylation reaction, have been solved in free form and
bound to its substrate pyruvate, product oxaloacetate, or inhibitor
2-ketobutyrate. The structure reveals a dimer of beta(8)alpha(8) barrels with an
active site cobalt ion coordinated by a carbamylated lysine, except in the
oxaloacetate complex in which the product's carboxylate group serves as a ligand
instead. 5S and human pyruvate carboxylase (PC), an enzyme crucial to
gluconeogenesis, catalyze similar reactions. A 5S-based homology model of the PC
carboxyltransferase domain indicates a conserved mechanism and explains the
molecular basis of mutations in lactic acidemia. PC disease mutations reproduced
in 5S result in a similar decrease in carboxyltransferase activity and crystal
structures with altered active sites.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5 Active sites of 5S complexes. The free 5S active site
is shown with its Lys[C]184 (ball-and-stick with gray carbon
atoms and bonds), cobalt ion (pink sphere), water ligands (red
spheres), and side chains of residues which interact with either
the cobalt ion or bound ligand. Superimposed are oxaloacetate
(yellow), pyruvate (pale pink), and 2-ketobutyrate (cyan)
ligands from their respective complexes.
|
 |
Figure 6.
Figure 6 Active sites of 5S mutants. (A) Composite 5S active
site showing the 5S-A59T crystal structure. The Thr59 side chain
(orange and red ball-and-stick) interacts with Gln26, which in
the complex structures forms hydrogen bonds with the bound
ligands. For reference, oxaloacetate (yellow, as bound in
5S-oxal) and residues with which it interacts (Arg22 and Gln26),
Met186 (gray, conformation in free 5S; pale pink, conformation
in 5S-oxal), cobalt ion (pink sphere), and carbamylated
Lys[C]184 are also shown. The orientation of this figure is
related to that of Figure 5 by  90°
rotation about the vertical axis combined with 90°
rotation about the vertical axis combined with  90°
rotation about the horizontal axis. (B) Composite 5S active site
showing the 5S-M186I crystal structure. The I186 side chain
(cyan ball-and-stick structure) would be expected to make close
contacts with substrate or product; oxaloacetate (yellow, as
bound in 5S-oxal) and Ala59 are shown for reference. 90°
rotation about the horizontal axis. (B) Composite 5S active site
showing the 5S-M186I crystal structure. The I186 side chain
(cyan ball-and-stick structure) would be expected to make close
contacts with substrate or product; oxaloacetate (yellow, as
bound in 5S-oxal) and Ala59 are shown for reference.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2004,
23,
3621-3631)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Falentin,
S.M.Deutsch,
G.Jan,
V.Loux,
A.Thierry,
S.Parayre,
M.B.Maillard,
J.Dherbécourt,
F.J.Cousin,
J.Jardin,
P.Siguier,
A.Couloux,
V.Barbe,
B.Vacherie,
P.Wincker,
J.F.Gibrat,
C.Gaillardin,
and
S.Lortal
(2010).
The complete genome of Propionibacterium freudenreichii CIRM-BIA1, a hardy actinobacterium with food and probiotic applications.
|
| |
PLoS One,
5,
e11748.
|
 |
|
|
|
|
 |
S.Duangpan,
S.Jitrapakdee,
A.Adina-Zada,
L.Byrne,
T.N.Zeczycki,
M.St Maurice,
W.W.Cleland,
J.C.Wallace,
and
P.V.Attwood
(2010).
Probing the catalytic roles of Arg548 and Gln552 in the carboxyl transferase domain of the Rhizobium etli pyruvate carboxylase by site-directed mutagenesis.
|
| |
Biochemistry,
49,
3296-3304.
|
 |
|
|
|
|
 |
T.Granjon,
O.Maniti,
Y.Auchli,
P.Dahinden,
R.Buchet,
O.Marcillat,
and
P.Dimroth
(2010).
Structure-function relations in oxaloacetate decarboxylase complex. Fluorescence and infrared approaches to monitor oxomalonate and Na(+) binding effect.
|
| |
PLoS One,
5,
e10935.
|
 |
|
|
|
|
 |
C.Y.Chou,
L.P.Yu,
and
L.Tong
(2009).
Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism.
|
| |
J Biol Chem,
284,
11690-11697.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.P.Yu,
S.Xiang,
G.Lasso,
D.Gil,
M.Valle,
and
L.Tong
(2009).
A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A.
|
| |
Structure,
17,
823-832.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.D.Townsend,
P.M.Holliday,
S.Fenyk,
K.C.Hess,
M.A.Gray,
D.R.Hodgson,
and
M.J.Cann
(2009).
Stimulation of Mammalian G-protein-responsive Adenylyl Cyclases by Carbon Dioxide.
|
| |
J Biol Chem,
284,
784-791.
|
 |
|
|
|
|
 |
T.N.Zeczycki,
M.St Maurice,
S.Jitrapakdee,
J.C.Wallace,
P.V.Attwood,
and
W.W.Cleland
(2009).
Insight into the carboxyl transferase domain mechanism of pyruvate carboxylase from Rhizobium etli.
|
| |
Biochemistry,
48,
4305-4313.
|
 |
|
|
|
|
 |
S.Jitrapakdee,
M.St Maurice,
I.Rayment,
W.W.Cleland,
J.C.Wallace,
and
P.V.Attwood
(2008).
Structure, mechanism and regulation of pyruvate carboxylase.
|
| |
Biochem J,
413,
369-387.
|
 |
|
|
|
|
 |
S.Xiang,
and
L.Tong
(2008).
Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction.
|
| |
Nat Struct Mol Biol,
15,
295-302.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Li,
C.H.Hagemeier,
H.Seedorf,
G.Gottschalk,
and
R.K.Thauer
(2007).
Re-citrate synthase from Clostridium kluyveri is phylogenetically related to homocitrate synthase and isopropylmalate synthase rather than to Si-citrate synthase.
|
| |
J Bacteriol,
189,
4299-4304.
|
 |
|
|
|
|
 |
J.D.Burman,
C.E.Stevenson,
R.G.Sawers,
and
D.M.Lawson
(2007).
The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family.
|
| |
BMC Struct Biol,
7,
30.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.St Maurice,
L.Reinhardt,
K.H.Surinya,
P.V.Attwood,
J.C.Wallace,
W.W.Cleland,
and
I.Rayment
(2007).
Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme.
|
| |
Science,
317,
1076-1079.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Z.Ozimek,
S.H.Klompmaker,
N.Visser,
M.Veenhuis,
and
I.J.van der Klei
(2007).
The transcarboxylase domain of pyruvate carboxylase is essential for assembly of the peroxisomal flavoenzyme alcohol oxidase.
|
| |
FEMS Yeast Res,
7,
1082-1092.
|
 |
|
|
|
|
 |
S.Friedmann,
B.E.Alber,
and
G.Fuchs
(2007).
Properties of R-citramalyl-coenzyme A lyase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus.
|
| |
J Bacteriol,
189,
2906-2914.
|
 |
|
|
|
|
 |
F.Forouhar,
M.Hussain,
R.Farid,
J.Benach,
M.Abashidze,
W.C.Edstrom,
S.M.Vorobiev,
R.Xiao,
T.B.Acton,
Z.Fu,
J.J.Kim,
H.M.Miziorko,
G.T.Montelione,
and
J.F.Hunt
(2006).
Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metalloenzymes cleaving carbon-carbon bonds.
|
| |
J Biol Chem,
281,
7533-7545.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.R.Carey
(2006).
Raman crystallography and other biochemical applications of Raman microscopy.
|
| |
Annu Rev Phys Chem,
57,
527-554.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

