
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 597 a.a.
597 a.a.
|
 |

|

|

|

|

|

|

|
 20 a.a.
20 a.a.
|
 |

|

|

|

|

|

|

|
 18 a.a.
18 a.a.
|
 |

|

|

|

|

|

|

|
 14 a.a.
14 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Inhibitor bound human angiotensin converting enzyme-related carboxypeptidase (ace2)
|
|
Structure:
|
 |
Angiotensin i converting enzyme 2. Chain: a. Fragment: extracellular domains. Synonym: angiotensin converting enzyme-like protein, angiotensin converting enzyme-related carboxypeptidase. Engineered: yes. Disordered segment of collectrin homology domain. Chain: b. Engineered: yes.
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: ace2. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108. Expression_system_cell_line: sf9.
|
|
Resolution:
|
 |
|
3.00Å
|
R-factor:
|
0.253
|
R-free:
|
0.337
|
|
|
Authors:
|
 |
P.Towler,B.Staker,S.G.Prasad,S.Menon,D.Ryan,J.Tang,T.Parsons, M.Fisher,D.Williams,N.A.Dales,M.A.Patane,M.W.Pantoliano
|
Key ref:

|
 |
P.Towler
et al.
(2004).
ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J Biol Chem,
279,
17996-18007.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
07-Oct-03
|
Release date:
|
03-Feb-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9BYF1
(ACE2_HUMAN) -
Angiotensin-converting enzyme 2 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
805 a.a.
597 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
No UniProt id for this chain
|
|

|

|
|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.3.4.17.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chain A:
E.C.3.4.17.23
- angiotensin-converting enzyme 2.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
angiotensin II + H2O = angiotensin-1-7 + L-phenylalanine
|
 |
 |
 |
 |
 |
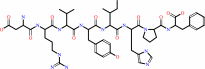 angiotensin II
angiotensin II
|
+
|
H2O
|
=
|
angiotensin-(1-7)
|
+
|
L-phenylalanine
Bound ligand (Het Group name = )
matches with 44.44% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:17996-18007
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
|
|
P.Towler,
B.Staker,
S.G.Prasad,
S.Menon,
J.Tang,
T.Parsons,
D.Ryan,
M.Fisher,
D.Williams,
N.A.Dales,
M.A.Patane,
M.W.Pantoliano.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The angiotensin-converting enzyme (ACE)-related carboxypeptidase, ACE2, is a
type I integral membrane protein of 805 amino acids that contains one HEXXH + E
zinc-binding consensus sequence. ACE2 has been implicated in the regulation of
heart function and also as a functional receptor for the coronavirus that causes
the severe acute respiratory syndrome (SARS). To gain further insights into this
enzyme, the first crystal structures of the native and inhibitor-bound forms of
the ACE2 extracellular domains were solved to 2.2- and 3.0-A resolution,
respectively. Comparison of these structures revealed a large
inhibitor-dependent hinge-bending movement of one catalytic subdomain relative
to the other ( approximately 16 degrees ) that brings important residues into
position for catalysis. The potent inhibitor MLN-4760
((S,S)-2-[1-carboxy-2-[3-(3,5-dichlorobenzyl)-3H-imidazol4-yl]-ethylamino]-4-methylpentanoic
acid) makes key binding interactions within the active site and offers insights
regarding the action of residues involved in catalysis and substrate
specificity. A few active site residue substitutions in ACE2 relative to ACE
appear to eliminate the S(2)' substrate-binding subsite and account for the
observed reactivity change from the peptidyl dipeptidase activity of ACE to the
carboxypeptidase activity of ACE2.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIG. 4. Superposition of the native and inhibitor-bound
ACE2 structures. A, the 409  -carbon atoms
corresponding to subdomain II of the native and inhibitor-bound
ACE2 structures were superimposed with an r.m.s. deviation of
1.41 Å. Native ACE2 is colored red, and inhibitor-bound
ACE2 is colored green. The zinc ion is shown as a yellow sphere,
and the inhibitor MLN-4760 is shown in a ball-and-stick
rendering with default atom coloring: gray, carbon; blue,
nitrogen; red, oxygen; green, chlorine. This view is looking
down the length of the active site cleft and is rotated 90°
from that shown in Fig. 3. This perspective illustrates the -carbon atoms
corresponding to subdomain II of the native and inhibitor-bound
ACE2 structures were superimposed with an r.m.s. deviation of
1.41 Å. Native ACE2 is colored red, and inhibitor-bound
ACE2 is colored green. The zinc ion is shown as a yellow sphere,
and the inhibitor MLN-4760 is shown in a ball-and-stick
rendering with default atom coloring: gray, carbon; blue,
nitrogen; red, oxygen; green, chlorine. This view is looking
down the length of the active site cleft and is rotated 90°
from that shown in Fig. 3. This perspective illustrates the  16°
hinge-bending movement of subdomain I relative to subdomain II
that occurs upon inhibitor binding to ACE2. B, shown is a
close-up view of the active sites of the superimposed native
(red) and inhibitor-bound (green) ACE2 structures. This is the
same superposition of subdomain II for both structures as
described for A. In this perspective, the residues of subdomain
I within the active site are shown to move upon inhibitor
binding relative to those in subdomain II. The inhibitor
MLN-4760 is shown in stick rendering with the same atom color
code as described for A. The average movement for residues near
the active site is 6-9 Å. The yellow spheres are the two
positions of the zinc atom in the native and inhibitor-bound
structures. This figure was prepared using MOE 2003.02 software
(Chemical Computing Group, Inc.). 16°
hinge-bending movement of subdomain I relative to subdomain II
that occurs upon inhibitor binding to ACE2. B, shown is a
close-up view of the active sites of the superimposed native
(red) and inhibitor-bound (green) ACE2 structures. This is the
same superposition of subdomain II for both structures as
described for A. In this perspective, the residues of subdomain
I within the active site are shown to move upon inhibitor
binding relative to those in subdomain II. The inhibitor
MLN-4760 is shown in stick rendering with the same atom color
code as described for A. The average movement for residues near
the active site is 6-9 Å. The yellow spheres are the two
positions of the zinc atom in the native and inhibitor-bound
structures. This figure was prepared using MOE 2003.02 software
(Chemical Computing Group, Inc.).
|
 |
Figure 6.
FIG. 6. Superposition of the ACE2 and tACE structures. A,
the  -carbon atoms in
lisinopril-bound tACE (13) were superimposed onto the equivalent
atoms in inhibitor-bound ACE2 (588 residues) with an r.m.s.
deviation of 1.80 Å. MLN-4760-bound ACE2 is magenta, and
lisinopril-bound tACE is green. MLN-4760 is shown bound to ACE2
with the same color code described in the legend to Fig. 4A.
Similarly, the zinc and chloride ions are shown as described in
the legend to Fig. 3. The orientation is the same as that shown
for native ACE2 in Fig. 3. Structures were superimposed using
MOE 2003.02 software. B, the 21 -carbon atoms in
lisinopril-bound tACE (13) were superimposed onto the equivalent
atoms in inhibitor-bound ACE2 (588 residues) with an r.m.s.
deviation of 1.80 Å. MLN-4760-bound ACE2 is magenta, and
lisinopril-bound tACE is green. MLN-4760 is shown bound to ACE2
with the same color code described in the legend to Fig. 4A.
Similarly, the zinc and chloride ions are shown as described in
the legend to Fig. 3. The orientation is the same as that shown
for native ACE2 in Fig. 3. Structures were superimposed using
MOE 2003.02 software. B, the 21  -carbon atoms at the
inhibitor-bound active site of ACE2 (residues 4.5 Å from
the inhibitor) were superimposed onto the equivalent atoms of
lisinopril-bound tACE (Protein Data Bank code 1O86 [PDB]
) with an r.m.s. deviation of 0.53 Å. The active site of
ACE2 and MLN-4760 are shown in default colors, with the
inhibitor displayed in stick rendering. Labels are for ACE2
residues only. The active site residues of tACE are shown in
orange, with the inhibitor lisinopril colored purple in stick
rendering. The zinc ion is shown as a yellow sphere, and the
second chloride ion of tACE (CL2) is shown as an orange sphere.
This chloride ion site does not exist in ACE2 due to the Glu398
substitution for Pro407 (see "Results and Discussion"). Other
important differences between ACE2 and tACE are as follows:
Arg273 versus Gln281, Phe^274 versus Thr282, and Tyr510 versus
Val518, respectively. -carbon atoms at the
inhibitor-bound active site of ACE2 (residues 4.5 Å from
the inhibitor) were superimposed onto the equivalent atoms of
lisinopril-bound tACE (Protein Data Bank code 1O86 [PDB]
) with an r.m.s. deviation of 0.53 Å. The active site of
ACE2 and MLN-4760 are shown in default colors, with the
inhibitor displayed in stick rendering. Labels are for ACE2
residues only. The active site residues of tACE are shown in
orange, with the inhibitor lisinopril colored purple in stick
rendering. The zinc ion is shown as a yellow sphere, and the
second chloride ion of tACE (CL2) is shown as an orange sphere.
This chloride ion site does not exist in ACE2 due to the Glu398
substitution for Pro407 (see "Results and Discussion"). Other
important differences between ACE2 and tACE are as follows:
Arg273 versus Gln281, Phe^274 versus Thr282, and Tyr510 versus
Val518, respectively.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
17996-18007)
copyright 2004.
|
|
| |
Figures were
selected
by the author.
|
|

|
| |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|

|
| |
One interesting feature of this zinc metallopeptidase is the large hinge-bending conformation change that occurs upon inhibitor (and presumably also substrate) binding. A similar conformational change is believed to occur for inhibitor and substrate binding to the closest homologs of ACE2: testicular and somatic ACE. These enzymes are all now believed to play critical roles in the renin angiotensin system for bood pressure homeostasis.
Another interesting feature of ACE2 is the discovery that it has been high jacked to serve as the functional receptor of the SARS coronovirus (Li et al. 2003, Nature 426, 450-454). However, inhibitors of ACE2 such as MLN-4760, shown bound to the active site in these crystal structures, do not block binding of the virus. The reason for this became apparent after publication of the structure of the SARS coronovirus spike protein bound to ACE2 (see Li, et al. 2005, SCIENCE 309, p1864-1868). This paper revealed that the virus binds to the N-terminal lobe of the peptidase, far removed from the peptidase catalytic center.
Michael W. Pantoliano
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Clayton,
I.Hanchapola,
N.Hausler,
S.Unabia,
R.A.Lew,
R.E.Widdop,
A.I.Smith,
P.Perlmutter,
and
M.I.Aguilar
(2011).
β-amino acid substitution to investigate the recognition of angiotensin II (AngII) by angiotensin converting enzyme 2 (ACE2).
|
| |
J Mol Recognit,
24,
235-244.
|
 |
|
|
|
|
 |
T.T.Nguyen,
S.C.Chang,
I.Evnouchidou,
I.A.York,
C.Zikos,
K.L.Rock,
A.L.Goldberg,
E.Stratikos,
and
L.J.Stern
(2011).
Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1.
|
| |
Nat Struct Mol Biol,
18,
604-613.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Kuba,
Y.Imai,
T.Ohto-Nakanishi,
and
J.M.Penninger
(2010).
Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters.
|
| |
Pharmacol Ther,
128,
119-128.
|
 |
|
|
|
|
 |
H.P.Jia,
D.C.Look,
P.Tan,
L.Shi,
M.Hickey,
L.Gakhar,
M.C.Chappell,
C.Wohlford-Lenane,
and
P.B.McCray
(2009).
Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia.
|
| |
Am J Physiol Lung Cell Mol Physiol,
297,
L84-L96.
|
 |
|
|
|
|
 |
K.Wu,
W.Li,
G.Peng,
and
F.Li
(2009).
Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor.
|
| |
Proc Natl Acad Sci U S A,
106,
19970-19974.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.C.Mathewson,
A.Bishop,
Y.Yao,
F.Kemp,
J.Ren,
H.Chen,
X.Xu,
B.Berkhout,
L.van der Hoek,
and
I.M.Jones
(2008).
Interaction of severe acute respiratory syndrome-coronavirus and NL63 coronavirus spike proteins with angiotensin converting enzyme-2.
|
| |
J Gen Virol,
89,
2741-2745.
|
 |
|
|
|
|
 |
A.J.Ferreira,
J.A.Hernández Prada,
D.A.Ostrov,
and
M.K.Raizada
(2008).
Cardiovascular protection by angiotensin-converting enzyme 2: a new paradigm.
|
| |
Future Cardiol,
4,
175-182.
|
 |
|
|
|
|
 |
C.A.Rushworth,
J.L.Guy,
and
A.J.Turner
(2008).
Residues affecting the chloride regulation and substrate selectivity of the angiotensin-converting enzymes (ACE and ACE2) identified by site-directed mutagenesis.
|
| |
FEBS J,
275,
6033-6042.
|
 |
|
|
|
|
 |
C.E.Isaza,
X.Zhong,
L.E.Rosas,
J.D.White,
R.P.Chen,
G.F.Liang,
S.I.Chan,
A.R.Satoskar,
and
M.K.Chan
(2008).
A proposed role for Leishmania major carboxypeptidase in peptide catabolism.
|
| |
Biochem Biophys Res Commun,
373,
25-29.
|
 |
|
|
|
|
 |
D.Batlle,
M.J.Soler,
and
J.Wysocki
(2008).
New aspects of the renin-angiotensin system: angiotensin-converting enzyme 2 - a potential target for treatment of hypertension and diabetic nephropathy.
|
| |
Curr Opin Nephrol Hypertens,
17,
250-257.
|
 |
|
|
|
|
 |
F.Li
(2008).
Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections.
|
| |
J Virol,
82,
6984-6991.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Wysocki,
F.R.González-Pacheco,
and
D.Batlle
(2008).
Angiotensin-converting enzyme 2: possible role in hypertension and kidney disease.
|
| |
Curr Hypertens Rep,
10,
70-77.
|
 |
|
|
|
|
 |
M.Han,
W.Yan,
Y.Huang,
H.Yao,
Z.Wang,
D.Xi,
W.Li,
Y.Zhou,
J.Hou,
X.Luo,
and
Q.Ning
(2008).
The nucleocapsid protein of SARS-CoV induces transcription of hfgl2 prothrombinase gene dependent on C/EBP alpha.
|
| |
J Biochem,
144,
51-62.
|
 |
|
|
|
|
 |
A.J.Trask,
and
C.M.Ferrario
(2007).
Angiotensin-(1-7): pharmacology and new perspectives in cardiovascular treatments.
|
| |
Cardiovasc Drug Rev,
25,
162-174.
|
 |
|
|
|
|
 |
M.F.Doobay,
L.S.Talman,
T.D.Obr,
X.Tian,
R.L.Davisson,
and
E.Lazartigues
(2007).
Differential expression of neuronal ACE2 in transgenic mice with overexpression of the brain renin-angiotensin system.
|
| |
Am J Physiol Regul Integr Comp Physiol,
292,
R373-R381.
|
 |
|
|
|
|
 |
M.Rella,
J.L.Elliot,
T.J.Revett,
J.Lanfear,
A.Phelan,
R.M.Jackson,
A.J.Turner,
and
N.M.Hooper
(2007).
Identification and characterisation of the angiotensin converting enzyme-3 (ACE3) gene: a novel mammalian homologue of ACE.
|
| |
BMC Genomics,
8,
194.
|
 |
|
|
|
|
 |
V.C.Cheng,
S.K.Lau,
P.C.Woo,
and
K.Y.Yuen
(2007).
Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection.
|
| |
Clin Microbiol Rev,
20,
660-694.
|
 |
|
|
|
|
 |
E.De Clercq
(2006).
Potential antivirals and antiviral strategies against SARS coronavirus infections.
|
| |
Expert Rev Anti Infect Ther,
4,
291-302.
|
 |
|
|
|
|
 |
J.M.Watermeyer,
B.T.Sewell,
S.L.Schwager,
R.Natesh,
H.R.Corradi,
K.R.Acharya,
and
E.D.Sturrock
(2006).
Structure of testis ACE glycosylation mutants and evidence for conserved domain movement.
|
| |
Biochemistry,
45,
12654-12663.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.S.Yeung,
G.A.Yamanaka,
and
N.A.Meanwell
(2006).
Severe acute respiratory syndrome coronavirus entry into host cells: Opportunities for therapeutic intervention.
|
| |
Med Res Rev,
26,
414-433.
|
 |
|
|
|
|
 |
R.J.Bingham,
V.Dive,
S.E.Phillips,
A.D.Shirras,
and
R.E.Isaac
(2006).
Structural diversity of angiotensin-converting enzyme.
|
| |
FEBS J,
273,
362-373.
|
 |
|
|
|
|
 |
S.Bhushan,
K.A.Johnson,
T.Eneqvist,
and
E.Glaser
(2006).
Proteolytic mechanism of a novel mitochondrial and chloroplastic PreP peptidasome.
|
| |
Biol Chem,
387,
1087-1090.
|
 |
|
|
|
|
 |
A.G.Tzakos,
and
I.P.Gerothanassis
(2005).
Domain-selective ligand-binding modes and atomic level pharmacophore refinement in angiotensin I converting enzyme (ACE) inhibitors.
|
| |
Chembiochem,
6,
1089-1103.
|
 |
|
|
|
|
 |
H.Hofmann,
K.Pyrc,
L.van der Hoek,
M.Geier,
B.Berkhout,
and
S.Pöhlmann
(2005).
Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry.
|
| |
Proc Natl Acad Sci U S A,
102,
7988-7993.
|
 |
|
|
|
|
 |
J.L.Guy,
R.M.Jackson,
H.A.Jensen,
N.M.Hooper,
and
A.J.Turner
(2005).
Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis.
|
| |
FEBS J,
272,
3512-3520.
|
 |
|
|
|
|
 |
L.W.Yang,
and
I.Bahar
(2005).
Coupling between catalytic site and collective dynamics: a requirement for mechanochemical activity of enzymes.
|
| |
Structure,
13,
893-904.
|
 |
|
|
|
|
 |
S.Chakraborti,
P.Prabakaran,
X.Xiao,
and
D.S.Dimitrov
(2005).
The SARS coronavirus S glycoprotein receptor binding domain: fine mapping and functional characterization.
|
| |
Virol J,
2,
73.
|
 |
|
|
|
|
 |
S.S.Wong,
and
K.Y.Yuen
(2005).
The severe acute respiratory syndrome (SARS).
|
| |
J Neurovirol,
11,
455-468.
|
 |
|
|
|
|
 |
W.Li,
C.Zhang,
J.Sui,
J.H.Kuhn,
M.J.Moore,
S.Luo,
S.K.Wong,
I.C.Huang,
K.Xu,
N.Vasilieva,
A.Murakami,
Y.He,
W.A.Marasco,
Y.Guan,
H.Choe,
and
M.Farzan
(2005).
Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2.
|
| |
EMBO J,
24,
1634-1643.
|
 |
|
|
|
|
 |
A.S.Galanis,
G.A.Spyroulias,
G.Pairas,
E.Manessi-Zoupa,
and
P.Cordopatis
(2004).
Solid-phase synthesis and conformational properties of angiotensin converting enzyme catalytic-site peptides: the basis for a structural study on the enzyme-substrate interaction.
|
| |
Biopolymers,
76,
512-526.
|
 |
|
|
|
|
 |
A.Tousignant,
and
J.N.Pelletier
(2004).
Protein motions promote catalysis.
|
| |
Chem Biol,
11,
1037-1042.
|
 |
|
|
|
|
 |
E.De Clercq
(2004).
Antivirals and antiviral strategies.
|
| |
Nat Rev Microbiol,
2,
704-720.
|
 |
|
|
|
|
 |
H.Hofmann,
and
S.Pöhlmann
(2004).
Cellular entry of the SARS coronavirus.
|
| |
Trends Microbiol,
12,
466-472.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|

| |

