 |
PDBsum entry 1qgo

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Metal binding protein
|
PDB id
|
|

|

|
1qgo
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.99.1.3
- sirohydrochlorin cobaltochelatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Corrin and Siroheme Biosynthesis (part 2)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
Co-sirohydrochlorin + 2 H+ = sirohydrochlorin + Co2+
|
|
2.
|
Co-precorrin-2 + 3 H+ = precorrin-2 + Co2+
|
|
 |
 |
 |
 |
 |
Co-sirohydrochlorin
|
+
|
2
×
H(+)
|
=
|
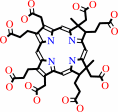 sirohydrochlorin
sirohydrochlorin
|
+
|
Co(2+)
|
|
 |
 |
 |
 |
 |
Co-precorrin-2
|
+
|
3
×
H(+)
|
=
|
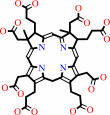 precorrin-2
precorrin-2
|
+
|
Co(2+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
38:10660-10669
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis.
|
|
H.L.Schubert,
E.Raux,
K.S.Wilson,
M.J.Warren.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Prosthetic groups such as heme, chlorophyll, and cobalamin (vitamin B(12)) are
characterized by their branched biosynthetic pathway and unique metal insertion
steps. The metal ion chelatases can be broadly classed either as single-subunit
ATP-independent enzymes, such as the anaerobic cobalt chelatase and the
protoporphyrin IX (PPIX) ferrochelatase, or as heterotrimeric, ATP-dependent
enzymes, such as the Mg chelatase involved in chlorophyll biosynthesis. The
X-ray structure of the anaerobic cobalt chelatase from Salmonella typhimurium,
CbiK, has been solved to 2.4 A resolution. Despite a lack of significant amino
acid sequence similarity, the protein structure is homologous to that of
Bacillus subtilis PPIX ferrochelatase. Both enzymes contain a histidine residue
previously identified as the metal ion ligand, but CbiK contains a second
histidine in place of the glutamic acid residue identified as a general base in
PPIX ferrochelatase. Site-directed mutagenesis has confirmed a role for this
histidine and a nearby glutamic acid in cobalt binding, modulating metal ion
specificity as well as catalytic efficiency. Contrary to the predicted
protoporphyrin binding site in PPIX ferrochelatase, the precorrin-2 binding site
in CbiK is clearly defined within a large horizontal cleft between the N- and
C-terminal domains. The structural similarity has implications for the
understanding of the evolution of this branched biosynthetic pathway.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.V.Romão,
D.Ladakis,
S.A.Lobo,
M.A.Carrondo,
A.A.Brindley,
E.Deery,
P.M.Matias,
R.W.Pickersgill,
L.M.Saraiva,
and
M.J.Warren
(2011).
Evolution in a family of chelatases facilitated by the introduction of active site asymmetry and protein oligomerization.
|
| |
Proc Natl Acad Sci U S A,
108,
97.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.C.Tripathy,
I.Sherameti,
and
R.Oelmüller
(2010).
Siroheme: an essential component for life on earth.
|
| |
Plant Signal Behav,
5,
14-20.
|
 |
|
|
|
|
 |
J.Lundqvist,
H.Elmlund,
R.P.Wulff,
L.Berglund,
D.Elmlund,
C.Emanuelsson,
H.Hebert,
R.D.Willows,
M.Hansson,
M.Lindahl,
and
S.Al-Karadaghi
(2010).
ATP-induced conformational dynamics in the AAA+ motor unit of magnesium chelatase.
|
| |
Structure,
18,
354-365.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.A.Hunter,
M.P.Sampson,
and
G.C.Ferreira
(2008).
Metal ion substrate inhibition of ferrochelatase.
|
| |
J Biol Chem,
283,
23685-23691.
|
 |
|
|
|
|
 |
H.L.Schubert,
R.S.Rose,
H.K.Leech,
A.A.Brindley,
C.P.Hill,
S.E.Rigby,
and
M.J.Warren
(2008).
Structure and function of SirC from Bacillus megaterium: a metal-binding precorrin-2 dehydrogenase.
|
| |
Biochem J,
415,
257-263.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Karlberg,
M.D.Hansson,
R.K.Yengo,
R.Johansson,
H.O.Thorvaldsen,
G.C.Ferreira,
M.Hansson,
and
S.Al-Karadaghi
(2008).
Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues.
|
| |
J Mol Biol,
378,
1074-1083.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.L.Holliday,
J.M.Thornton,
A.Marquet,
A.G.Smith,
F.Rébeillé,
R.Mendel,
H.L.Schubert,
A.D.Lawrence,
and
M.J.Warren
(2007).
Evolution of enzymes and pathways for the biosynthesis of cofactors.
|
| |
Nat Prod Rep,
24,
972-987.
|
 |
|
|
|
|
 |
R.R.Mendel,
A.G.Smith,
A.Marquet,
and
M.J.Warren
(2007).
Metal and cofactor insertion.
|
| |
Nat Prod Rep,
24,
963-971.
|
 |
|
|
|
|
 |
A.Andreeva,
and
A.G.Murzin
(2006).
Evolution of protein fold in the presence of functional constraints.
|
| |
Curr Opin Struct Biol,
16,
399-408.
|
 |
|
|
|
|
 |
J.Yin,
L.X.Xu,
M.M.Cherney,
E.Raux-Deery,
A.A.Bindley,
A.Savchenko,
J.R.Walker,
M.E.Cuff,
M.J.Warren,
and
M.N.James
(2006).
Crystal structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus.
|
| |
J Struct Funct Genomics,
7,
37-50.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.D.Hansson,
M.Lindstam,
and
M.Hansson
(2006).
Crosstalk between metal ions in Bacillus subtilis ferrochelatase.
|
| |
J Biol Inorg Chem,
11,
325-333.
|
 |
|
|
|
|
 |
E.Raux-Deery,
H.K.Leech,
K.A.Nakrieko,
K.J.McLean,
A.W.Munro,
P.Heathcote,
S.E.Rigby,
A.G.Smith,
and
M.J.Warren
(2005).
Identification and characterization of the terminal enzyme of siroheme biosynthesis from Arabidopsis thaliana: a plastid-located sirohydrochlorin ferrochelatase containing a 2FE-2S center.
|
| |
J Biol Chem,
280,
4713-4721.
|
 |
|
|
|
|
 |
A.A.Brindley,
E.Raux,
H.K.Leech,
H.L.Schubert,
and
M.J.Warren
(2003).
A story of chelatase evolution: identification and characterization of a small 13-15-kDa "ancestral" cobaltochelatase (CbiXS) in the archaea.
|
| |
J Biol Chem,
278,
22388-22395.
|
 |
|
|
|
|
 |
D.A.Rodionov,
A.G.Vitreschak,
A.A.Mironov,
and
M.S.Gelfand
(2003).
Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes.
|
| |
J Biol Chem,
278,
41148-41159.
|
 |
|
|
|
|
 |
H.K.Leech,
E.Raux,
K.J.McLean,
A.W.Munro,
N.J.Robinson,
G.P.Borrelly,
M.Malten,
D.Jahn,
S.E.Rigby,
P.Heathcote,
and
M.J.Warren
(2003).
Characterization of the cobaltochelatase CbiXL: evidence for a 4Fe-4S center housed within an MXCXXC motif.
|
| |
J Biol Chem,
278,
41900-41907.
|
 |
|
|
|
|
 |
J.E.Cornah,
M.J.Terry,
and
A.G.Smith
(2003).
Green or red: what stops the traffic in the tetrapyrrole pathway?
|
| |
Trends Plant Sci,
8,
224-230.
|
 |
|
|
|
|
 |
M.E.Stroupe,
H.K.Leech,
D.S.Daniels,
M.J.Warren,
and
E.D.Getzoff
(2003).
CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis.
|
| |
Nat Struct Biol,
10,
1064-1073.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.A.Whittaker,
and
R.O.Hynes
(2002).
Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere.
|
| |
Mol Biol Cell,
13,
3369-3387.
|
 |
|
|
|
|
 |
H.L.Schubert,
E.Raux,
A.A.Brindley,
H.K.Leech,
K.S.Wilson,
C.P.Hill,
and
M.J.Warren
(2002).
The structure of Saccharomyces cerevisiae Met8p, a bifunctional dehydrogenase and ferrochelatase.
|
| |
EMBO J,
21,
2068-2075.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.P.Keller,
P.M.Smith,
J.Benach,
D.Christendat,
G.T.deTitta,
and
J.F.Hunt
(2002).
The crystal structure of MT0146/CbiT suggests that the putative precorrin-8w decarboxylase is a methyltransferase.
|
| |
Structure,
10,
1475-1487.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.G.Dashper,
A.Hendtlass,
N.Slakeski,
C.Jackson,
K.J.Cross,
L.Brownfield,
R.Hamilton,
I.Barr,
and
E.C.Reynolds
(2000).
Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis.
|
| |
J Bacteriol,
182,
6456-6462.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

