 |
PDBsum entry 1ldb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase(choh(d)-NAD(a))
|
PDB id
|
|

|

|
1ldb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.27
- L-lactate dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-lactate + NAD+ = pyruvate + NADH + H+
|
 |
 |
 |
 |
 |
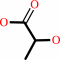 (S)-lactate
(S)-lactate
|
+
|
 NAD(+)
NAD(+)
|
=
|
 pyruvate
pyruvate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Proteins
7:74-92
(1990)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure determination and refinement of Bacillus stearothermophilus lactate dehydrogenase.
|
|
K.Piontek,
P.Chakrabarti,
H.P.Schär,
M.G.Rossmann,
H.Zuber.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Structures have been determined of Bacillus stearothermophilus "apo" and holo
lactate dehydrogenase. The holo-enzyme had been co-crystallized with the
activator fructose 1,6-bisphosphate. The "apo" lactate dehydrogenase structure
was solved by use of the known apo-M4 dogfish lactate dehydrogenase molecule as
a starting model. Phases were refined and extended from 4 A to 3 A resolution by
means of the noncrystallographic molecular 222 symmetry. The R-factor was
reduced to 28.7%, using 2.8 A resolution data, in a restrained least-squares
refinement in which the molecular symmetry was imposed as a constraint. A low
occupancy of coenzyme was found in each of the four subunits of the
"apo"-enzyme. Further refinement proceeded with the isomorphous holo-enzyme from
Bacillus stearothermophilus. After removing the noncrystallographic constraints,
the R-factor dropped from 30.3% to a final value of 26.0% with a 0.019 A and 1.7
degrees r.m.s. deviation from idealized bond lengths and angles, respectively.
Two sulfate ions per subunit were included in the final model of the
"apo"-form--one at the substrate binding site and one close to the molecular
P-axis near the location of the fructose 1,6-bisphosphate activator. The final
model of the holo-enzyme incorporated two sulfate ions per subunit, one at the
substrate binding site and another close to the R-axis. One nicotinamide adenine
dinucleotide coenzyme molecule per subunit and two fructose 1,6-bisphosphate
molecules per tetramer were also included. The phosphate positions of fructose
1,6-bisphosphate are close to the sulfate ion near the P-axis in the "apo"
model. This structure represents the first reported refined model of an
allosteric activated lactate dehydrogenase. The structure of the activated
holo-enzyme showed far greater similarity to the ternary complex of dogfish M4
lactate dehydrogenase with nicotinamide adenine dinucleotide and oxamate than to
apo-M4 dogfish lactate dehydrogenase. The conformations of nicotinamide adenine
dinucleotide and fructose 1,6-bisphosphate were also analyzed.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Uchikoba,
S.Fushinobu,
T.Wakagi,
M.Konno,
H.Taguchi,
and
H.Matsuzawa
(2002).
Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
|
| |
Proteins,
46,
206-214.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Read,
V.J.Winter,
C.M.Eszes,
R.B.Sessions,
and
R.L.Brady
(2001).
Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
|
| |
Proteins,
43,
175-185.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Fieulaine,
S.Morera,
S.Poncet,
V.Monedero,
V.Gueguen-Chaignon,
A.Galinier,
J.Janin,
J.Deutscher,
and
S.Nessler
(2001).
X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain.
|
| |
EMBO J,
20,
3917-3927.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.I.Lee,
C.Chang,
S.J.Cho,
G.W.Han,
Y.G.Yu,
S.H.Eom,
and
S.W.Suh
(2000).
Lactate dehydrogenase from the hyperthermophilic archaeon Methanococcus jannaschii: overexpression, crystallization and preliminary X-ray analysis.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
81-83.
|
 |
|
|
|
|
 |
S.Kochhar,
V.S.Lamzin,
A.Razeto,
M.Delley,
H.Hottinger,
and
J.E.Germond
(2000).
Roles of his205, his296, his303 and Asp259 in catalysis by NAD+-specific D-lactate dehydrogenase.
|
| |
Eur J Biochem,
267,
1633-1639.
|
 |
|
|
|
|
 |
J.A.Read,
K.W.Wilkinson,
R.Tranter,
R.B.Sessions,
and
R.L.Brady
(1999).
Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
|
| |
J Biol Chem,
274,
10213-10218.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Rabinovich,
H.Rozenberg,
and
Z.Shakked
(1998).
Molecular replacement: the revival of the molecular Fourier transform method.
|
| |
Acta Crystallogr D Biol Crystallogr,
54,
1336-1342.
|
 |
|
|
|
|
 |
K.Savijoki,
and
A.Palva
(1997).
Molecular genetic characterization of the L-lactate dehydrogenase gene (ldhL) of Lactobacillus helveticus and biochemical characterization of the enzyme.
|
| |
Appl Environ Microbiol,
63,
2850-2856.
|
 |
|
|
|
|
 |
M.Xie,
J.Seravalli,
W.P.Huskey,
K.B.Schowen,
and
R.L.Schowen
(1994).
Solvent isotope effects and the nature of electrophilic catalysis in the action of the lactate dehydrogenase of Bacillus stearothermophilus.
|
| |
Bioorg Med Chem,
2,
691-695.
|
 |
|
|
|
|
 |
S.Iwata,
K.Kamata,
S.Yoshida,
T.Minowa,
and
T.Ohta
(1994).
T and R states in the crystals of bacterial L-lactate dehydrogenase reveal the mechanism for allosteric control.
|
| |
Nat Struct Biol,
1,
176-185.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.Livnah,
E.A.Bayer,
M.Wilchek,
and
J.L.Sussman
(1993).
Three-dimensional structures of avidin and the avidin-biotin complex.
|
| |
Proc Natl Acad Sci U S A,
90,
5076-5080.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Ostendorp,
W.Liebl,
H.Schurig,
and
R.Jaenicke
(1993).
The L-lactate dehydrogenase gene of the hyperthermophilic bacterium Thermotoga maritima cloned by complementation in Escherichia coli.
|
| |
Eur J Biochem,
216,
709-715.
|
 |
|
|
|
|
 |
K.Huang,
R.Kodandapani,
H.Kallwass,
J.K.Hogan,
W.Parris,
J.D.Friesen,
M.Gold,
J.B.Jones,
and
M.N.James
(1992).
Crystallization and preliminary X-ray diffraction studies of two mutants of lactate dehydrogenase from Bacillus stearothermophilus.
|
| |
Proteins,
13,
158-161.
|
 |
|
|
|
|
 |
S.Kochhar,
H.Hottinger,
N.Chuard,
P.G.Taylor,
T.Atkinson,
M.D.Scawen,
and
D.J.Nicholls
(1992).
Cloning and overexpression of Lactobacillus helveticus D-lactate dehydrogenase gene in Escherichia coli.
|
| |
Eur J Biochem,
208,
799-805.
|
 |
|
|
|
|
 |
D.Ghosh,
C.M.Weeks,
P.Grochulski,
W.L.Duax,
M.Erman,
R.L.Rimsay,
and
J.C.Orr
(1991).
Three-dimensional structure of holo 3 alpha,20 beta-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family.
|
| |
Proc Natl Acad Sci U S A,
88,
10064-10068.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.C.Lawrence
(1991).
The application of the molecular replacement method to the de novo determination of protein structure.
|
| |
Q Rev Biophys,
24,
399-424.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

