 |
PDBsum entry 1kd0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.1.2
- methylaspartate ammonia-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2S,3S)-3-methyl-L-aspartate = mesaconate + NH4+
|
 |
 |
 |
 |
 |
(2S,3S)-3-methyl-L-aspartate
|
=
|
 mesaconate
mesaconate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cob(II)alamin
|
 |
 |
 |
 |
 |
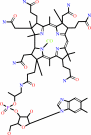 Cob(II)alamin
Cob(II)alamin
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:8306-8311
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
|
|
M.Asuncion,
W.Blankenfeldt,
J.N.Barlow,
D.Gani,
J.H.Naismith.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Methylaspartate ammonia-lyase (3-methylaspartase, MAL; EC ) catalyzes the
reversible anti elimination of ammonia from L-threo-(2S,3S)-3-methylaspartic
acid to give mesaconic acid. This reaction lies on the main catabolic pathway
for glutamate in Clostridium tetanomorphum. MAL requires monovalent and divalent
cation cofactors for full catalytic activity. The enzyme has attracted interest
because of its potential use as a biocatalyst. The structure of C. tetanomorphum
MAL has been solved to 1.9-A resolution by the single-wavelength anomalous
diffraction method. A divalent metal ion complex of the protein has also been
determined. MAL is a homodimer with each monomer consisting of two domains. One
is an alpha/beta-barrel, and the other smaller domain is mainly beta-strands.
The smaller domain partially occludes the C terminus of the barrel and forms a
large cleft. The structure identifies MAL as belonging to the enolase
superfamily of enzymes. The metal ion site is located in a large cleft between
the domains. Potential active site residues have been identified based on a
combination of their proximity to a metal ion site, molecular modeling, and
sequence homology. In common with all members of the enolase superfamily, the
carboxylic acid of the substrate is co-ordinated by the metal ions, and a proton
adjacent to a carboxylic acid group of the substrate is abstracted by a base. In
MAL, it appears that Lys(331) removes the alpha-proton of methylaspartic acid.
This motif is the defining mechanistic characteristic of the enolase superfamily
of which all have a common fold. The degree of structural conservation is
remarkable given only four residues are absolutely conserved.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. The reaction catalyzed by methylaspartase. The
reverse reaction is both stereo- and regiospecific. Harnessing
this reaction for organic synthesis would provide a new route to
homochiral aspartic acids.
|
 |
Figure 4.
Fig. 4. The model of MAL with its substrate at the active
site. The residues that bind the metal ion are shown along with
those inferred to be the catalytic base (Lys331) and substrate
recognition (Arg80). His194, which may be important in
deprotonating the other diastereomer is also shown. MASP,
methylaspartate. The original position of the side chains of
Arg80 and Lys331 are shown in gray.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
8306-8311)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Raj,
B.Weiner,
V.P.Veetil,
C.R.Reis,
W.J.Quax,
D.B.Janssen,
B.L.Feringa,
and
G.J.Poelarends
(2009).
Alteration of the diastereoselectivity of 3-methylaspartate ammonia lyase by using structure-based mutagenesis.
|
| |
Chembiochem,
10,
2236-2245.
|
 |
|
|
|
|
 |
N.Sapay,
Y.Guermeur,
and
G.Deléage
(2006).
Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier.
|
| |
BMC Bioinformatics,
7,
255.
|
 |
|
|
|
|
 |
W.Buckel,
and
B.T.Golding
(2006).
Radical enzymes in anaerobes.
|
| |
Annu Rev Microbiol,
60,
27-49.
|
 |
|
|
|
|
 |
L.Poppe,
and
J.Rétey
(2005).
Friedel-Crafts-type mechanism for the enzymatic elimination of ammonia from histidine and phenylalanine.
|
| |
Angew Chem Int Ed Engl,
44,
3668-3688.
|
 |
|
|
|
|
 |
E.C.Meng,
B.J.Polacco,
and
P.C.Babbitt
(2004).
Superfamily active site templates.
|
| |
Proteins,
55,
962-976.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

