 |
PDBsum entry 1jhq

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.21
- nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Corrin Biosynthesis (part 8)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5,6-dimethylbenzimidazole + nicotinate beta-D-ribonucleotide = alpha- ribazole 5'-phosphate + nicotinate + H+
|
 |
 |
 |
 |
 |
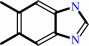 5,6-dimethylbenzimidazole
5,6-dimethylbenzimidazole
|
+
|
nicotinate beta-D-ribonucleotide
|
=
|
alpha- ribazole 5'-phosphate
Bound ligand (Het Group name = )
matches with 84.62% similarity
|
+
|
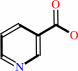 nicotinate
nicotinate
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
276:37612-37620
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
|
|
C.G.Cheong,
J.C.Escalante-Semerena,
I.Rayment.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Nicotinate mononucleotide (NaMN):5,6-dimethylbenzimidazole
phosphoribosyltransferase (CobT) from Salmonella enterica plays a central role
in the synthesis of alpha-ribazole, a key component of the lower ligand of
cobalamin. Surprisingly, CobT can phosphoribosylate a wide range of aromatic
substrates, giving rise to a wide variety of lower ligands in cobamides. To
understand the molecular basis for this lack of substrate specificity, the x-ray
structures of CobT complexed with adenine, 5-methylbenzimidazole,
5-methoxybenzimidazole, p-cresol, and phenol were determined. Furthermore,
adenine, 5-methylbenzimidazole, 5-methoxybenzimidazole, and 2-hydroxypurine were
observed to react with NaMN within the crystal lattice and undergo the
phosphoribosyl transfer reaction to form product. Significantly, the
stereochemistries of all products are identical to those found in vivo.
Interestingly, p-cresol and phenol, which are the lower ligand in Sporomusa
ovata, bound to CobT but did not react with NaMN. This study provides a
structural explanation for how CobT can phosphoribosylate most of the commonly
observed lower ligands found in cobamides with the exception of the phenolic
lower ligands observed in S. ovata. This is accomplished with minor
conformational changes in the side chains that constitute the
5,6-dimethylbenzimidazole binding site. These investigations are consistent with
the implication that the nature of the lower ligand is controlled by metabolic
factors rather by the specificity of the phosphoribosyltransferase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Fig. 4. Difference electron density for the ligands or
products of the reaction with NaMN complexed with CobT. Shown
are adenine (A),  -adenosine
monophosphate and nicotinate (B), 5-methylbenzimidazole (C),
N1-(5-phospho- -adenosine
monophosphate and nicotinate (B), 5-methylbenzimidazole (C),
N1-(5-phospho-  -ribosyl)-5-methylbenzimidazole
and nicotinate (D), 5-methoxybenzimidazole (E), and
N1-(5-phospho- -ribosyl)-5-methylbenzimidazole
and nicotinate (D), 5-methoxybenzimidazole (E), and
N1-(5-phospho-  -ribosyl)-5-methoxybenzimidazole
and nicotinate (F). Coefficients of the form F[o] -ribosyl)-5-methoxybenzimidazole
and nicotinate (F). Coefficients of the form F[o]  F[c] were
utilized where the ligand was excluded from the phase
calculation. The maps were contoured at the level of 1 F[c] were
utilized where the ligand was excluded from the phase
calculation. The maps were contoured at the level of 1  . .
|
 |
Figure 6.
Fig. 6. Difference electron density for the reaction
product for 2-hydroxypurine and NaMN , N1-(5-phospho-  -ribosyl)-2-hydroxypurine,
and nicotinate (A), p-cresol (B), p-cresol and nicotinate (C),
phenol (D), and phenol and nicotinate complexed with CobT (E),
respectively. Coefficients of the form F[o] -ribosyl)-2-hydroxypurine,
and nicotinate (A), p-cresol (B), p-cresol and nicotinate (C),
phenol (D), and phenol and nicotinate complexed with CobT (E),
respectively. Coefficients of the form F[o]  F[c] were
utilized, where the ligand was excluded from the phase
calculation. The maps were contoured at the level of 1 F[c] were
utilized, where the ligand was excluded from the phase
calculation. The maps were contoured at the level of 1  . .
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2001,
276,
37612-37620)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.R.Claas,
J.R.Parrish,
L.A.Maggio-Hall,
and
J.C.Escalante-Semerena
(2010).
Functional analysis of the nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) enzyme, involved in the late steps of coenzyme B12 biosynthesis in Salmonella enterica.
|
| |
J Bacteriol,
192,
145-154.
|
 |
|
|
|
|
 |
M.E.Taga,
and
G.C.Walker
(2008).
Pseudo-B12 joins the cofactor family.
|
| |
J Bacteriol,
190,
1157-1159.
|
 |
|
|
|
|
 |
C.L.Zayas,
and
J.C.Escalante-Semerena
(2007).
Reassessment of the late steps of coenzyme B12 synthesis in Salmonella enterica: evidence that dephosphorylation of adenosylcobalamin-5'-phosphate by the CobC phosphatase is the last step of the pathway.
|
| |
J Bacteriol,
189,
2210-2218.
|
 |
|
|
|
|
 |
J.C.Escalante-Semerena
(2007).
Conversion of cobinamide into adenosylcobamide in bacteria and archaea.
|
| |
J Bacteriol,
189,
4555-4560.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

