 |
PDBsum entry 1eeh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.9
- UDP-N-acetylmuramoyl-L-alanine--D-glutamate ligase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Peptidoglycan Biosynthesis (Part 1)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
UDP-N-acetyl-alpha-D-muramoyl-L-alanine + D-glutamate + ATP = UDP-N- acetyl-alpha-D-muramoyl-L-alanyl-D-glutamate + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
UDP-N-acetyl-alpha-D-muramoyl-L-alanine
|
+
|
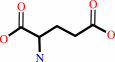 D-glutamate
D-glutamate
|
+
|
 ATP
ATP
|
=
|
UDP-N- acetyl-alpha-D-muramoyl-L-alanyl-D-glutamate
Bound ligand (Het Group name = )
matches with 43.40% similarity
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
301:1257-1266
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
"Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
|
|
J.A.Bertrand,
E.Fanchon,
L.Martin,
L.Chantalat,
G.Auger,
D.Blanot,
J.van Heijenoort,
O.Dideberg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
UDP-N-acetylmuramoyl-l-alanine:d-glutamate (MurD) ligase catalyses the addition
of d-glutamate to the nucleotide precursor UDP-N-acetylmuramoyl-l-alanine (UMA).
The crystal structures of Escherichia coli in the substrate-free form and MurD
complexed with UMA have been determined at 2.4 A and 1.88 A resolution,
respectively. The MurD structure comprises three domains each of a topology
reminiscent of nucleotide-binding folds. In the two structures the C-terminal
domain undergoes a large rigid-body rotation away from the N-terminal and
central domains. These two "open" structures were compared with the
four published "closed" structures of MurD. In addition the comparison
reveals which regions are affected by the binding of UMA, ATP and d-Glu. Also we
compare and discuss two structurally characterized enzymes which belong to the
same ligase superfamily: MurD and folylpolyglutamate synthetase (FGS). The
analysis allows the identification of key residues involved in the reaction
mechanism of FGS. The determination of the two "open" conformation
structures represents a new step towards the complete elucidation of the
enzymatic mechanism of the MurD ligase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5. Stereo view showing the "closed" form model of
FGS (black) superimposed on the central and C-terminal domains
of MurD.UMA.ADP.Mg2+ (green). The Image -Ala of UMA is shown in
red and ADP in blue.
|
 |
Figure 6.
Figure 6. Stereo view of the active-site region of the
"closed" conformation of FGS. Residues that play a role in the
binding of either ADP.Mg2+ and/or the terminal carboxylate group
of UMA are labelled. The carboxylate group of UMA is shown in
orange, ADP in deep blue, water molecules in red and Mg2+ in
black. The blue lines show strand b6, the P-loop and the
beginning of helix a6.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2000,
301,
1257-1266)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.Sosič,
H.Barreteau,
M.Simčič,
R.Sink,
J.Cesar,
A.Zega,
S.G.Grdadolnik,
C.Contreras-Martel,
A.Dessen,
A.Amoroso,
B.Joris,
D.Blanot,
and
S.Gobec
(2011).
Second-generation sulfonamide inhibitors of d-glutamic acid-adding enzyme: Activity optimisation with conformationally rigid analogues of d-glutamic acid.
|
| |
Eur J Med Chem,
46,
2880-2894.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Tomasić,
N.Zidar,
A.Kovac,
S.Turk,
M.Simcic,
D.Blanot,
M.Müller-Premru,
M.Filipic,
S.G.Grdadolnik,
A.Zega,
M.Anderluh,
S.Gobec,
D.Kikelj,
and
L.Peterlin Masic
(2010).
5-Benzylidenethiazolidin-4-ones as multitarget inhibitors of bacterial Mur ligases.
|
| |
ChemMedChem,
5,
286-295.
|
 |
|
|
|
|
 |
A.Perdih,
M.Hodoscek,
and
T.Solmajer
(2009).
MurD ligase from E. coli: Tetrahedral intermediate formation study by hybrid quantum mechanical/molecular mechanical replica path method.
|
| |
Proteins,
74,
744-759.
|
 |
|
|
|
|
 |
A.Perdih,
U.Bren,
and
T.Solmajer
(2009).
Binding free energy calculations of N-sulphonyl-glutamic acid inhibitors of MurD ligase.
|
| |
J Mol Model,
15,
983-996.
|
 |
|
|
|
|
 |
C.Paradis-Bleau,
A.Lloyd,
F.Sanschagrin,
H.Maaroufi,
T.Clarke,
A.Blewett,
C.Dowson,
D.I.Roper,
T.D.Bugg,
and
R.C.Levesque
(2009).
Pseudomonas aeruginosa MurE amide ligase: enzyme kinetics and peptide inhibitor.
|
| |
Biochem J,
421,
263-272.
|
 |
|
|
|
|
 |
C.Paradis-Bleau,
A.Lloyd,
F.Sanschagrin,
T.Clarke,
A.Blewett,
T.D.Bugg,
and
R.C.Levesque
(2008).
Phage display-derived inhibitor of the essential cell wall biosynthesis enzyme MurF.
|
| |
BMC Biochem,
9,
33.
|
 |
|
|
|
|
 |
H.Barreteau,
A.Kovac,
A.Boniface,
M.Sova,
S.Gobec,
and
D.Blanot
(2008).
Cytoplasmic steps of peptidoglycan biosynthesis.
|
| |
FEMS Microbiol Rev,
32,
168-207.
|
 |
|
|
|
|
 |
T.Bratkovic,
M.Lunder,
U.Urleb,
and
B.Strukelj
(2008).
Peptide inhibitors of MurD and MurE, essential enzymes of bacterial cell wall biosynthesis.
|
| |
J Basic Microbiol,
48,
202-206.
|
 |
|
|
|
|
 |
A.Perdih,
M.Kotnik,
M.Hodoscek,
and
T.Solmajer
(2007).
Targeted molecular dynamics simulation studies of binding and conformational changes in E. coli MurD.
|
| |
Proteins,
68,
243-254.
|
 |
|
|
|
|
 |
G.Füser,
and
A.Steinbüchel
(2007).
Analysis of genome sequences for genes of cyanophycin metabolism: identifying putative cyanophycin metabolizing prokaryotes.
|
| |
Macromol Biosci,
7,
278-296.
|
 |
|
|
|
|
 |
K.L.Longenecker,
G.F.Stamper,
P.J.Hajduk,
E.H.Fry,
C.G.Jakob,
J.E.Harlan,
R.Edalji,
D.M.Bartley,
K.A.Walter,
L.R.Solomon,
T.F.Holzman,
Y.G.Gu,
C.G.Lerner,
B.A.Beutel,
and
V.S.Stoll
(2005).
Structure of MurF from Streptococcus pneumoniae co-crystallized with a small molecule inhibitor exhibits interdomain closure.
|
| |
Protein Sci,
14,
3039-3047.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Mathieu,
G.Debousker,
S.Vincent,
F.Viviani,
N.Bamas-Jacques,
and
V.Mikol
(2005).
Escherichia coli FolC structure reveals an unexpected dihydrofolate binding site providing an attractive target for anti-microbial therapy.
|
| |
J Biol Chem,
280,
18916-18922.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.D.Mol,
A.Brooun,
D.R.Dougan,
M.T.Hilgers,
L.W.Tari,
R.A.Wijnands,
M.W.Knuth,
D.E.McRee,
and
R.V.Swanson
(2003).
Crystal structures of active fully assembled substrate- and product-bound complexes of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) from Haemophilus influenzae.
|
| |
J Bacteriol,
185,
4152-4162.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Li,
H.Xu,
D.E.Graham,
and
R.H.White
(2003).
Glutathione synthetase homologs encode alpha-L-glutamate ligases for methanogenic coenzyme F420 and tetrahydrosarcinapterin biosyntheses.
|
| |
Proc Natl Acad Sci U S A,
100,
9785-9790.
|
 |
|
|
|
|
 |
T.Deva,
K.D.Pryor,
B.Leiting,
E.N.Baker,
and
C.A.Smith
(2003).
Purification, crystallization and preliminary X-ray analysis of Escherichia coli UDP-N-acetylmuramoyl:L-alanine ligase (MurC).
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1510-1513.
|
 |
|
|
|
|
 |
D.W.Green
(2002).
The bacterial cell wall as a source of antibacterial targets.
|
| |
Expert Opin Ther Targets,
6,
1.
|
 |
|
|
|
|
 |
Y.Urushibata,
S.Tokuyama,
and
Y.Tahara
(2002).
Characterization of the Bacillus subtilis ywsC gene, involved in gamma-polyglutamic acid production.
|
| |
J Bacteriol,
184,
337-343.
|
 |
|
|
|
|
 |
S.Dementin,
A.Bouhss,
G.Auger,
C.Parquet,
D.Mengin-Lecreulx,
O.Dideberg,
J.van Heijenoort,
and
D.Blanot
(2001).
Evidence of a functional requirement for a carbamoylated lysine residue in MurD, MurE and MurF synthetases as established by chemical rescue experiments.
|
| |
Eur J Biochem,
268,
5800-5807.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

