 |
PDBsum entry 1e4l

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the inactive mutant monocot (maize zmglu1) beta- glucosidase zm glu191asp
|
|
Structure:
|
 |
Beta-glucosidase, chloroplastic. Chain: a, b. Synonym: beta-d-glucoside glucohydrolase, cellobiase, gentiobiase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Zea mays. Maize. Organism_taxid: 4577. Strain: cv. Mutin. Tissue: coleoptile. Organelle: chloroplast. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_cell: plys s cells.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.207
|
R-free:
|
0.244
|
|
|
Authors:
|
 |
M.Czjzek,M.Cicek,D.R.Bevan,B.Henrissat,A.Esen
|
Key ref:

|
 |
M.Czjzek
et al.
(2000).
The mechanism of substrate (aglycone) specificity in beta -glucosidases is revealed by crystal structures of mutant maize beta -glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes.
Proc Natl Acad Sci U S A,
97,
13555-13560.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
10-Jul-00
|
Release date:
|
11-Dec-00
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P49235
(HGGL1_MAIZE) -
4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside beta-D-glucosidase 1, chloroplastic from Zea mays
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
566 a.a.
490 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.2.1.182
- 4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
DIMBOA beta-D-glucoside + H2O = DIMBOA + D-glucose
|
|
2.
|
DIBOA beta-D-glucoside + H2O = DIBOA + D-glucose
|
|
 |
 |
 |
 |
 |
DIMBOA beta-D-glucoside
|
+
|
H2O
|
=
|
DIMBOA
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
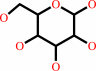 D-glucose
D-glucose
|
|
 |
 |
 |
 |
 |
DIBOA beta-D-glucoside
|
+
|
H2O
|
=
|
DIBOA
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
 D-glucose
D-glucose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.2.1.21
- beta-glucosidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Hydrolysis of terminal, non-reducing beta-D-glucose residues with release of beta-D-glucose.
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
97:13555-13560
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
The mechanism of substrate (aglycone) specificity in beta -glucosidases is revealed by crystal structures of mutant maize beta -glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes.
|
|
M.Czjzek,
M.Cicek,
V.Zamboni,
D.R.Bevan,
B.Henrissat,
A.Esen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The mechanism and the site of substrate (i.e., aglycone) recognition and
specificity were investigated in maize beta-glucosidase (Glu1) by x-ray
crystallography by using crystals of a catalytically inactive mutant (Glu1E191D)
in complex with the natural substrate
2-O-beta-d-glucopyranosyl-4-hydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOAGlc),
the free aglycone DIMBOA, and competitive inhibitor
para-hydroxy-S-mandelonitrile beta-glucoside (dhurrin). The structures of these
complexes and of the free enzyme were solved at 2.1-, 2.1-, 2.0-, and 2.2-A
resolution, respectively. The structural data from the complexes allowed us to
visualize an intact substrate, free aglycone, or a competitive inhibitor in the
slot-like active site of a beta-glucosidase. These data show that the aglycone
moiety of the substrate is sandwiched between W378 on one side and F198, F205,
and F466 on the other. Thus, specific conformations of these four hydrophobic
amino acids and the shape of the aglycone-binding site they form determine
aglycone recognition and substrate specificity in Glu1. In addition to these
four residues, A467 interacts with the 7-methoxy group of DIMBOA. All residues
but W378 are variable among beta-glucosidases that differ in substrate
specificity, supporting the conclusion that these sites are the basis of
aglycone recognition and binding (i.e., substrate specificity) in
beta-glucosidases. The data also provide a plausible explanation for the
competitive binding of dhurrin to maize beta-glucosidases with high affinity
without being hydrolyzed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Structure of the ligands and the active site of
Glu1E191D. (A) The natural substrate DIMBOAGlc (Left), the
aglycone DIMBOA (Center), and the competitive inhibitor dhurrin
(Right). (B) Ribbon diagram of the structure of the maize  -glucosidase
Glu1 and its inactive Glu1E191D mutant, showing the catalytic
residues E191 (D191 in the mutant) and E406 (red), four residues
(F198, F205, W378, and F466) forming the aglycone-binding pocket
(blue), and two other residues (A467 and Y473) that are probably
important for aglycone recognition (yellow). Different colors
and the color transitions in -glucosidase
Glu1 and its inactive Glu1E191D mutant, showing the catalytic
residues E191 (D191 in the mutant) and E406 (red), four residues
(F198, F205, W378, and F466) forming the aglycone-binding pocket
(blue), and two other residues (A467 and Y473) that are probably
important for aglycone recognition (yellow). Different colors
and the color transitions in  -helices
and -helices
and  -strands
trace the polypeptide backbone in the barrel-shaped
three-dimensional structure from the N terminus (dark blue) to
the C terminus (dark red) direction. The figure was produced
with MOLSCRIPT (35) and RASTER3D (36). (C) Electrostatic surface
representation of the active site region of Glu1E191D showing
positively charged regions in blue, negatively charged regions
in red, and neutral regions in white. The slot-like active site,
measuring 23 Å × 7.1 Å at the entrance,
contains the natural substrate DIMBOAGlc in compact
representation with standard atom-type colors. In this view,
only the aglycone moiety is visible in its binding site as
glucose is hidden below aglycone. C was produced with GRASP (37). -strands
trace the polypeptide backbone in the barrel-shaped
three-dimensional structure from the N terminus (dark blue) to
the C terminus (dark red) direction. The figure was produced
with MOLSCRIPT (35) and RASTER3D (36). (C) Electrostatic surface
representation of the active site region of Glu1E191D showing
positively charged regions in blue, negatively charged regions
in red, and neutral regions in white. The slot-like active site,
measuring 23 Å × 7.1 Å at the entrance,
contains the natural substrate DIMBOAGlc in compact
representation with standard atom-type colors. In this view,
only the aglycone moiety is visible in its binding site as
glucose is hidden below aglycone. C was produced with GRASP (37).
|
 |
Figure 3.
Fig. 3. Aglycone recognition and binding in  -glycosidases
as revealed by DIMBOAGlc-, DIMBOA-, and dhurrin-Glu1E191D
inactive mutant complexes. (A) Closeup view of the active site
of Glu1, showing the catalytic glutamates E191 and E406 (red),
the four residues (F198, F205, W378, and F466) forming the
aglycone-binding pocket (light blue), and the additional
residues (A467 and Y473) that are probably important for
aglycone recognition (light green). (B) Glu1E191D with bound
DIMBOAGlc. The glycone moiety is in blue, whereas the aglycone
is in atom-type colors. The bulky aryl group is sandwiched
between W378 on one side and F198, F205, and F466 on the other.
(C) Same as B but with bound DIMBOA, showing a slightly
different orientation than DIMBOA in DIMBOAGlc, which is
constrained by the glycosidic linkage. (D) Same as B but with
bound dhurrin. The aglycone moiety of the inhibitor dhurrin is
in the same position as the aglycone of the natural substrate.
Figs. 3. and 4 were produced with TURBO-FRODO (38). -glycosidases
as revealed by DIMBOAGlc-, DIMBOA-, and dhurrin-Glu1E191D
inactive mutant complexes. (A) Closeup view of the active site
of Glu1, showing the catalytic glutamates E191 and E406 (red),
the four residues (F198, F205, W378, and F466) forming the
aglycone-binding pocket (light blue), and the additional
residues (A467 and Y473) that are probably important for
aglycone recognition (light green). (B) Glu1E191D with bound
DIMBOAGlc. The glycone moiety is in blue, whereas the aglycone
is in atom-type colors. The bulky aryl group is sandwiched
between W378 on one side and F198, F205, and F466 on the other.
(C) Same as B but with bound DIMBOA, showing a slightly
different orientation than DIMBOA in DIMBOAGlc, which is
constrained by the glycosidic linkage. (D) Same as B but with
bound dhurrin. The aglycone moiety of the inhibitor dhurrin is
in the same position as the aglycone of the natural substrate.
Figs. 3. and 4 were produced with TURBO-FRODO (38).
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.R.Ketudat Cairns,
and
A.Esen
(2010).
β-Glucosidases.
|
| |
Cell Mol Life Sci,
67,
3389-3405.
|
 |
|
|
|
|
 |
V.Lombard,
T.Bernard,
C.Rancurel,
H.Brumer,
P.M.Coutinho,
and
B.Henrissat
(2010).
A hierarchical classification of polysaccharide lyases for glycogenomics.
|
| |
Biochem J,
432,
437-444.
|
 |
|
|
|
|
 |
A.D.Hill,
and
P.J.Reilly
(2008).
A Gibbs free energy correlation for automated docking of carbohydrates.
|
| |
J Comput Chem,
29,
1131-1141.
|
 |
|
|
|
|
 |
A.D.Hill,
and
P.J.Reilly
(2008).
Computational analysis of glycoside hydrolase family 1 specificities.
|
| |
Biopolymers,
89,
1021-1031.
|
 |
|
|
|
|
 |
L.M.Mendonça,
and
S.R.Marana
(2008).
The role in the substrate specificity and catalysis of residues forming the substrate aglycone-binding site of a beta-glycosidase.
|
| |
FEBS J,
275,
2536-2547.
|
 |
|
|
|
|
 |
R.Dopitová,
P.Mazura,
L.Janda,
R.Chaloupková,
P.Jerábek,
J.Damborský,
T.Filipi,
N.S.Kiran,
and
B.Brzobohatý
(2008).
Functional analysis of the aglycone-binding site of the maize beta-glucosidase Zm-p60.1.
|
| |
FEBS J,
275,
6123-6135.
|
 |
|
|
|
|
 |
J.Stöckigt,
and
S.Panjikar
(2007).
Structural biology in plant natural product biosynthesis--architecture of enzymes from monoterpenoid indole and tropane alkaloid biosynthesis.
|
| |
Nat Prod Rep,
24,
1382-1400.
|
 |
|
|
|
|
 |
R.Opassiri,
B.Pomthong,
T.Onkoksoong,
T.Akiyama,
A.Esen,
and
J.R.Ketudat Cairns
(2006).
Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 beta-glucosidase.
|
| |
BMC Plant Biol,
6,
33.
|
 |
|
|
|
|
 |
W.Chuenchor,
S.Pengthaisong,
J.Yuvaniyama,
R.Opassiri,
J.Svasti,
and
J.R.Ketudat Cairns
(2006).
Purification, crystallization and preliminary X-ray analysis of rice BGlu1 beta-glucosidase with and without 2-deoxy-2-fluoro-beta-D-glucoside.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
798-801.
|
 |
|
|
|
|
 |
J.Allouch,
W.Helbert,
B.Henrissat,
and
M.Czjzek
(2004).
Parallel substrate binding sites in a beta-agarase suggest a novel mode of action on double-helical agarose.
|
| |
Structure,
12,
623-632.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Xu,
L.Escamilla-Treviño,
L.Zeng,
M.Lalgondar,
D.Bevan,
B.Winkel,
A.Mohamed,
C.L.Cheng,
M.C.Shih,
J.Poulton,
and
A.Esen
(2004).
Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1.
|
| |
Plant Mol Biol,
55,
343-367.
|
 |
|
|
|
|
 |
A.Laederach,
and
P.J.Reilly
(2003).
Specific empirical free energy function for automated docking of carbohydrates to proteins.
|
| |
J Comput Chem,
24,
1748-1757.
|
 |
|
|
|
|
 |
B.Cobucci-Ponzano,
M.Moracci,
B.Di Lauro,
M.Ciaramella,
R.D'Avino,
and
M.Rossi
(2002).
Ionic network at the C-terminus of the beta-glycosidase from the hyperthermophilic archaeon Sulfolobus solfataricus: Functional role in the quaternary structure thermal stabilization.
|
| |
Proteins,
48,
98.
|
 |
|
|
|
|
 |
Y.Bhatia,
S.Mishra,
and
V.S.Bisaria
(2002).
Microbial beta-glucosidases: cloning, properties, and applications.
|
| |
Crit Rev Biotechnol,
22,
375-407.
|
 |
|
|
|
|
 |
Y.Bourne,
and
B.Henrissat
(2001).
Glycoside hydrolases and glycosyltransferases: families and functional modules.
|
| |
Curr Opin Struct Biol,
11,
593-600.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

