 |
PDBsum entry 1dht

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1dht
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.51
- 3(or 17)beta-hydroxysteroid dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
testosterone + NAD+ = androst-4-ene-3,17-dione + NADH + H+
|
|
2.
|
testosterone + NADP+ = androst-4-ene-3,17-dione + NADPH + H+
|
|
 |
 |
 |
 |
 |
 testosterone
testosterone
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
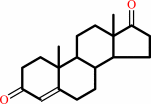 androst-4-ene-3,17-dione
androst-4-ene-3,17-dione
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 testosterone
testosterone
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 androst-4-ene-3,17-dione
androst-4-ene-3,17-dione
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.62
- 17beta-estradiol 17-dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
17beta-estradiol + NAD+ = estrone + NADH + H+
|
|
2.
|
17beta-estradiol + NADP+ = estrone + NADPH + H+
|
|
 |
 |
 |
 |
 |
17beta-estradiol
|
+
|
 NAD(+)
NAD(+)
|
=
|
estrone
Bound ligand (Het Group name = )
matches with 95.24% similarity
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
17beta-estradiol
|
+
|
 NADP(+)
NADP(+)
|
=
|
estrone
Bound ligand (Het Group name = )
matches with 95.24% similarity
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
275:1105-1111
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Dehydroepiandrosterone and dihydrotestosterone recognition by human estrogenic 17beta-hydroxysteroid dehydrogenase. C-18/c-19 steroid discrimination and enzyme-induced strain.
|
|
Q.Han,
R.L.Campbell,
A.Gangloff,
Y.W.Huang,
S.X.Lin.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Steroid hormones share a very similar structure, but they behave distinctly. We
present structures of human estrogenic 17beta-hydroxysteroid dehydrogenase
(17beta-HSD1) complexes with dehydroepiandrosterone (DHEA) and
dihydrotestosterone (DHT), providing the first pictures to date of DHEA and DHT
bound to a protein. Comparisons of these structures with that of the enzyme
complexed with the most potent estrogen, estradiol, revealed the structural
basis and general model for sex hormone recognition and discrimination. Although
the binding cavity is almost entirely composed of hydrophobic residues that can
make only nonspecific interactions, the arrangement of residues is highly
complementary to that of the estrogenic substrate. Relatively small changes in
the shape of the steroid hormone can significantly affect the binding affinity
and specificity. The K(m) of estrone is more than 1000-fold lower than that of
DHEA and the K(m) of estradiol is about 10 times lower than that of DHT. The
structures suggest that Leu-149 is the primary contributor to the discrimination
of C-19 steroids and estrogens by 17beta-HSD1. The critical role of Leu-149 has
been well confirmed by site-directed mutagenesis experiments, as the Leu-149 -->
Val variant showed a significantly decreased K(m) for C-19 steroids while losing
discrimination between estrogens and C-19 steroids. The electron density of DHEA
also revealed a distortion of its 17-ketone toward a beta-oriented form, which
approaches the transition-state conformation for DHEA reduction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Chemical structures of cyclo-pentenophenanthrene
ring, estrane ring, androstane ring, pregnane ring, DHEA,
progesterone, estradiol, and DHT.
|
 |
Figure 6.
Fig. 6. A side-by-side comparison of D-ring
conformations. The normal model of DHEA (right) is the
energy-minimized structure, which is very similar to the small
molecule crystal structure of DHEA (21). The D-ring conformation
of the model of 5-androstene-3,17-diol, the product of the
reduction of DHEA (left), was made using Insight II by referring
to the conformation of E[2]. The conformation of O-17 and the
D-ring of DHEA changes from  -oriented
toward an -oriented
toward an  -oriented
position during the reduction reaction. The strained model of
DHEA is also shown (center). -oriented
position during the reduction reaction. The strained model of
DHEA is also shown (center).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2000,
275,
1105-1111)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Negri,
M.Recanatini,
and
R.W.Hartmann
(2010).
Insights in 17beta-HSD1 enzyme kinetics and ligand binding by dynamic motion investigation.
|
| |
PLoS One,
5,
e12026.
|
 |
|
|
|
|
 |
S.Karkola,
A.Lilienkampf,
and
K.Wähälä
(2008).
A 3D QSAR model of 17beta-HSD1 inhibitors based on a thieno[2,3-d]pyrimidin-4(3H)-one core applying molecular dynamics simulations and ligand-protein docking.
|
| |
ChemMedChem,
3,
461-472.
|
 |
|
|
|
|
 |
M.Zhou,
W.Qiu,
H.J.Chang,
A.Gangloff,
and
S.X.Lin
(2002).
Purification, crystallization and preliminary X-ray diffraction results of human 17beta-hydroxysteroid dehydrogenase type 5.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
1048-1050.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

