 |
PDBsum entry 1d3v

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.3.1
- arginase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Urea Cycle and Arginine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-arginine + H2O = urea + L-ornithine
|
 |
 |
 |
 |
 |
 L-arginine
L-arginine
|
+
|
H2O
|
=
|
urea
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
+
|
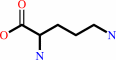 L-ornithine
L-ornithine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Biol
6:1043-1047
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Arginase-boronic acid complex highlights a physiological role in erectile function.
|
|
J.D.Cox,
N.N.Kim,
A.M.Traish,
D.W.Christianson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the complex between the binuclear manganese
metalloenzyme arginase and the boronic acid analog of L-arginine,
2(S)-amino-6-boronohexanoic acid (ABH), has been determined at 1.7 A resolution
from a crystal perfectly twinned by hemihedry. ABH binds as the tetrahedral
boronate anion, with one hydroxyl oxygen symmetrically bridging the binuclear
manganese cluster and a second hydroxyl oxygen coordinating to Mn2+A. This
binding mode mimics the transition state of a metal-activated hydroxide
mechanism. This transition state structure differs from that occurring in NO
biosynthesis, thereby explaining why ABH does not inhibit NO synthase. We also
show that arginase activity is present in the penis. Accordingly, the tight
binding and specificity of ABH allows us to probe the physiological role of
arginase in modulating the NO-dependent smooth muscle relaxation required for
erection. Strikingly, ABH causes significant enhancement of nonadrenergic,
noncholinergic nerve-mediated relaxation of penile corpus cavernosum smooth
muscle, suggesting that arginase inhibition sustains L-arginine concentrations
for NO synthase activity. Therefore, human penile arginase is a potential target
for therapeutic intervention in the treatment of erectile dysfunction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. L-Arginine catabolism. a, Structure-based mechanism
of arginase^16, in which metal-activated hydroxide ion attacks
the substrate guanidinium group to form a tetrahedral
intermediate (for clarity, only the side chain atoms of
substrate L-arginine are shown). Proton transfer mediated by Asp
128 facilitates collapse of this intermediate to form products
L-ornithine and urea. Following product dissociation, a
nucleophilic metal-bridging hydroxide ion is regenerated from a
metal-bridging water by proton transfer to bulk solvent. His 141
may function as a proton shuttle as indicated. b, Reciprocal
coordination of arginase and nitric oxide pathways; note that N^
 -hydroxy-L-arginine
is an intermediate in the NO synthase reaction. c, The arginase
inhibitor 2(S)-amino-6-boronohexanoic acid (ABH) is an isostere
of L-arginine. -hydroxy-L-arginine
is an intermediate in the NO synthase reaction. c, The arginase
inhibitor 2(S)-amino-6-boronohexanoic acid (ABH) is an isostere
of L-arginine.
|
 |
Figure 2.
Figure 2. Arginase−ABH complex. a, Omit electron density
map of ABH in the arginase active site averaged over the two
monomers in the asymmetric unit and averaged over the two twin
domains A and B as described in the text. The map is contoured
at 7.7  and
selected active site residues are indicated. Atoms are
color-coded as follows: C = yellow, O = red, N = blue, B = pale
green; water molecules appear as red spheres. This figure was
generated with BOBSCRIPT and Raster3D^34, ^35. b, Summary of
arginase−ABH interactions; manganese coordination interactions
are designated by green dashed lines, and hydrogen bonds are
indicated by black dashed lines. c, Stabilization of the
tetrahedral intermediate (and flanking transition states) in the
arginase mechanism based on the binding mode of ABH. and
selected active site residues are indicated. Atoms are
color-coded as follows: C = yellow, O = red, N = blue, B = pale
green; water molecules appear as red spheres. This figure was
generated with BOBSCRIPT and Raster3D^34, ^35. b, Summary of
arginase−ABH interactions; manganese coordination interactions
are designated by green dashed lines, and hydrogen bonds are
indicated by black dashed lines. c, Stabilization of the
tetrahedral intermediate (and flanking transition states) in the
arginase mechanism based on the binding mode of ABH.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Struct Biol
(1999,
6,
1043-1047)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Schade,
J.Kotthaus,
and
B.Clement
(2010).
Modulating the NO generating system from a medicinal chemistry perspective: current trends and therapeutic options in cardiovascular disease.
|
| |
Pharmacol Ther,
126,
279-300.
|
 |
|
|
|
|
 |
H.A.Toque,
M.J.Romero,
R.C.Tostes,
A.Shatanawi,
S.Chandra,
Z.N.Carneiro,
E.W.Inscho,
R.C.Webb,
R.B.Caldwell,
and
R.W.Caldwell
(2010).
p38 Mitogen-activated protein kinase (MAPK) increases arginase activity and contributes to endothelial dysfunction in corpora cavernosa from angiotensin-II-treated mice.
|
| |
J Sex Med,
7,
3857-3867.
|
 |
|
|
|
|
 |
L.Di Costanzo,
M.Ilies,
K.J.Thorn,
and
D.W.Christianson
(2010).
Inhibition of human arginase I by substrate and product analogues.
|
| |
Arch Biochem Biophys,
496,
101-108.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Y.Shishova,
L.Di Costanzo,
F.A.Emig,
D.E.Ash,
and
D.W.Christianson
(2009).
Probing the specificity determinants of amino acid recognition by arginase.
|
| |
Biochemistry,
48,
121-131.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.J.Ahn,
H.K.Chung,
C.H.Lee,
K.K.Kang,
and
B.O.Ahn
(2009).
Increased expression of the nitric oxide synthase gene and protein in corpus cavernosum by repeated dosing of udenafil in a rat model of chemical diabetogenesis.
|
| |
Asian J Androl,
11,
435-442.
|
 |
|
|
|
|
 |
M.Leopoldini,
N.Russo,
and
M.Toscano
(2009).
Determination of the catalytic pathway of a manganese arginase enzyme through density functional investigation.
|
| |
Chemistry,
15,
8026-8036.
|
 |
|
|
|
|
 |
D.Mavri-Damelin,
L.H.Damelin,
S.Eaton,
M.Rees,
C.Selden,
and
H.J.Hodgson
(2008).
Cells for bioartificial liver devices: the human hepatoma-derived cell line C3A produces urea but does not detoxify ammonia.
|
| |
Biotechnol Bioeng,
99,
644-651.
|
 |
|
|
|
|
 |
D.P.Dowling,
L.Di Costanzo,
H.A.Gennadios,
and
D.W.Christianson
(2008).
Evolution of the arginase fold and functional diversity.
|
| |
Cell Mol Life Sci,
65,
2039-2055.
|
 |
|
|
|
|
 |
H.Masuda
(2008).
Significance of nitric oxide and its modulation mechanisms by endogenous nitric oxide synthase inhibitors and arginase in the micturition disorders and erectile dysfunction.
|
| |
Int J Urol,
15,
128-134.
|
 |
|
|
|
|
 |
L.Santhanam,
D.W.Christianson,
D.Nyhan,
and
D.E.Berkowitz
(2008).
Arginase and vascular aging.
|
| |
J Appl Physiol,
105,
1632-1642.
|
 |
|
|
|
|
 |
B.Musicki,
and
A.L.Burnett
(2007).
Endothelial dysfunction in diabetic erectile dysfunction.
|
| |
Int J Impot Res,
19,
129-138.
|
 |
|
|
|
|
 |
L.Di Costanzo,
M.E.Pique,
and
D.W.Christianson
(2007).
Crystal structure of human arginase I complexed with thiosemicarbazide reveals an unusual thiocarbonyl mu-sulfide ligand in the binuclear manganese cluster.
|
| |
J Am Chem Soc,
129,
6388-6389.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Di Costanzo,
M.Moulin,
M.Haertlein,
F.Meilleur,
and
D.W.Christianson
(2007).
Expression, purification, assay, and crystal structure of perdeuterated human arginase I.
|
| |
Arch Biochem Biophys,
465,
82-89.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ghasemi,
H.Sadeghipour,
and
A.R.Dehpour
(2007).
Anandamide improves the impaired nitric oxide-mediated neurogenic relaxation of the corpus cavernosum in diabetic rats: involvement of cannabinoid CB1 and vanilloid VR1 receptors.
|
| |
BJU Int,
100,
1385-1390.
|
 |
|
|
|
|
 |
H.Maarsingh,
J.Leusink,
I.S.Bos,
J.Zaagsma,
and
H.Meurs
(2006).
Arginase strongly impairs neuronal nitric oxide-mediated airway smooth muscle relaxation in allergic asthma.
|
| |
Respir Res,
7,
6.
|
 |
|
|
|
|
 |
J.Steppan,
S.Ryoo,
K.H.Schuleri,
C.Gregg,
R.K.Hasan,
A.R.White,
L.J.Bugaj,
M.Khan,
L.Santhanam,
D.Nyhan,
A.A.Shoukas,
J.M.Hare,
and
D.E.Berkowitz
(2006).
Arginase modulates myocardial contractility by a nitric oxide synthase 1-dependent mechanism.
|
| |
Proc Natl Acad Sci U S A,
103,
4759-4764.
|
 |
|
|
|
|
 |
L.A.Holowatz,
C.S.Thompson,
and
W.L.Kenney
(2006).
L-Arginine supplementation or arginase inhibition augments reflex cutaneous vasodilatation in aged human skin.
|
| |
J Physiol,
574,
573-581.
|
 |
|
|
|
|
 |
R.Alarcón,
M.S.Orellana,
B.Neira,
E.Uribe,
J.R.García,
and
N.Carvajal
(2006).
Mutational analysis of substrate recognition by human arginase type I--agmatinase activity of the N130D variant.
|
| |
FEBS J,
273,
5625-5631.
|
 |
|
|
|
|
 |
R.C.Hillig,
and
L.Renault
(2006).
Detecting and overcoming hemihedral twinning during the MIR structure determination of Rna1p.
|
| |
Acta Crystallogr D Biol Crystallogr,
62,
750-765.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Maarsingh,
M.A.Tio,
J.Zaagsma,
and
H.Meurs
(2005).
Arginase attenuates inhibitory nonadrenergic noncholinergic nerve-induced nitric oxide generation and airway smooth muscle relaxation.
|
| |
Respir Res,
6,
23.
|
 |
|
|
|
|
 |
L.Di Costanzo,
G.Sabio,
A.Mora,
P.C.Rodriguez,
A.C.Ochoa,
F.Centeno,
and
D.W.Christianson
(2005).
Crystal structure of human arginase I at 1.29-A resolution and exploration of inhibition in the immune response.
|
| |
Proc Natl Acad Sci U S A,
102,
13058-13063.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.López,
R.Alarcón,
M.S.Orellana,
P.Enríquez,
E.Uribe,
J.Martínez,
and
N.Carvajal
(2005).
Insights into the interaction of human arginase II with substrate and manganese ions by site-directed mutagenesis and kinetic studies. Alteration of substrate specificity by replacement of Asn149 with Asp.
|
| |
FEBS J,
272,
4540-4548.
|
 |
|
|
|
|
 |
H.J.Ahn,
K.H.Kim,
J.Lee,
J.Y.Ha,
H.H.Lee,
D.Kim,
H.J.Yoon,
A.R.Kwon,
and
S.W.Suh
(2004).
Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily.
|
| |
J Biol Chem,
279,
50505-50513.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.Yang,
X.Gao,
and
B.Wang
(2003).
Boronic acid compounds as potential pharmaceutical agents.
|
| |
Med Res Rev,
23,
346-368.
|
 |
|
|
|
|
 |
H.Meurs,
S.McKay,
H.Maarsingh,
M.A.Hamer,
L.Macic,
N.Molendijk,
and
J.Zaagsma
(2002).
Increased arginase activity underlies allergen-induced deficiency of cNOS-derived nitric oxide and airway hyperresponsiveness.
|
| |
Br J Pharmacol,
136,
391-398.
|
 |
|
|
|
|
 |
R.K.Johansson,
M.Poljakovic,
K.E.Andersson,
and
K.Persson
(2002).
Expression of nitric oxide synthase in bladder smooth muscle cells: regulation by cytokines and L-arginine.
|
| |
J Urol,
168,
2280-2285.
|
 |
|
|
|
|
 |
S.M.Morris
(2002).
Regulation of enzymes of the urea cycle and arginine metabolism.
|
| |
Annu Rev Nutr,
22,
87.
|
 |
|
|
|
|
 |
D.M.Colleluori,
and
D.E.Ash
(2001).
Classical and slow-binding inhibitors of human type II arginase.
|
| |
Biochemistry,
40,
9356-9362.
|
 |
|
|
|
|
 |
N.N.Kim,
J.D.Cox,
R.F.Baggio,
F.A.Emig,
S.K.Mistry,
S.L.Harper,
D.W.Speicher,
S.M.Morris,
D.E.Ash,
A.Traish,
and
D.W.Christianson
(2001).
Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum.
|
| |
Biochemistry,
40,
2678-2688.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Moali,
M.Brollo,
J.Custot,
M.A.Sari,
J.L.Boucher,
D.J.Stuehr,
and
D.Mansuy
(2000).
Recognition of alpha-amino acids bearing various C=NOH functions by nitric oxide synthase and arginase involves very different structural determinants.
|
| |
Biochemistry,
39,
8208-8218.
|
 |
|
|
|
|
 |
H.Meurs,
M.A.Hamer,
S.Pethe,
S.Vadon-Le Goff,
J.L.Boucher,
and
J.Zaagsma
(2000).
Modulation of cholinergic airway reactivity and nitric oxide production by endogenous arginase activity.
|
| |
Br J Pharmacol,
130,
1793-1798.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

