 |
PDBsum entry 1cw2

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.4.2.1.20
- tryptophan synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate + L-serine = D-glyceraldehyde 3-phosphate + L-tryptophan + H2O
|
 |
 |
 |
 |
 |
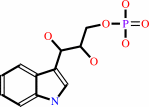 (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
 L-serine
L-serine
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
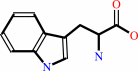 L-tryptophan
L-tryptophan
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
38:12665-12674
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
|
|
A.Sachpatzidis,
C.Dealwis,
J.B.Lubetsky,
P.H.Liang,
K.S.Anderson,
E.Lolis.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In an effort to use a structure-based approach for the design of new herbicides,
the crystal structures of complexes of tryptophan synthase with a series of
phosphonate enzyme inhibitors were determined at 2.3 A or higher resolution.
These inhibitors were designed to mimic the transition state formed during the
alpha-reaction of the enzyme and, as expected, have affinities much greater than
that of the natural substrate indole-3-glycerol phosphate or its nonhydrolyzable
analogue indole propanol phosphate (IPP). These inhibitors are ortho-substituted
arylthioalkylphosphonate derivatives that have an sp(3)-hybridized sulfur atom,
designed to mimic the putative tetrahedral transition state at the C3 atom of
the indole, and lack the C2 atom to allow for higher conformational flexibility.
Overall, the inhibitors bind in a fashion similar to that of IPP. Glu-49 and
Phe-212 are the two active site residues whose conformation changes upon
inhibitor binding. A very short hydrogen bond between a phosphonate oxygen and
the Ser-235 hydroxyl oxygen may be responsible for stabilization of the
enzyme-inhibitor complexes. Implications for the mechanism of catalysis as well
as directions for more potent inhibitors are discussed.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Nishio,
K.Ogasahara,
Y.Morimoto,
T.Tsukihara,
S.J.Lee,
and
K.Yutani
(2010).
Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding.
|
| |
FEBS J,
277,
2157-2170.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Raboni,
S.Bettati,
and
A.Mozzarelli
(2009).
Tryptophan synthase: a mine for enzymologists.
|
| |
Cell Mol Life Sci,
66,
2391-2403.
|
 |
|
|
|
|
 |
M.F.Dunn,
D.Niks,
H.Ngo,
T.R.Barends,
and
I.Schlichting
(2008).
Tryptophan synthase: the workings of a channeling nanomachine.
|
| |
Trends Biochem Sci,
33,
254-264.
|
 |
|
|
|
|
 |
M.Weyand,
I.Schlichting,
A.Marabotti,
and
A.Mozzarelli
(2002).
Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
|
| |
J Biol Chem,
277,
10647-10652.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Weyand,
I.Schlichting,
P.Herde,
A.Marabotti,
and
A.Mozzarelli
(2002).
Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
|
| |
J Biol Chem,
277,
10653-10660.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.W.Miles
(2001).
Tryptophan synthase: a multienzyme complex with an intramolecular tunnel.
|
| |
Chem Rec,
1,
140-151.
|
 |
|
|
|
|
 |
E.Weber-Ban,
O.Hur,
C.Bagwell,
U.Banik,
L.H.Yang,
E.W.Miles,
and
M.F.Dunn
(2001).
Investigation of allosteric linkages in the regulation of tryptophan synthase: the roles of salt bridges and monovalent cations probed by site-directed mutation, optical spectroscopy, and kinetics.
|
| |
Biochemistry,
40,
3497-3511.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

